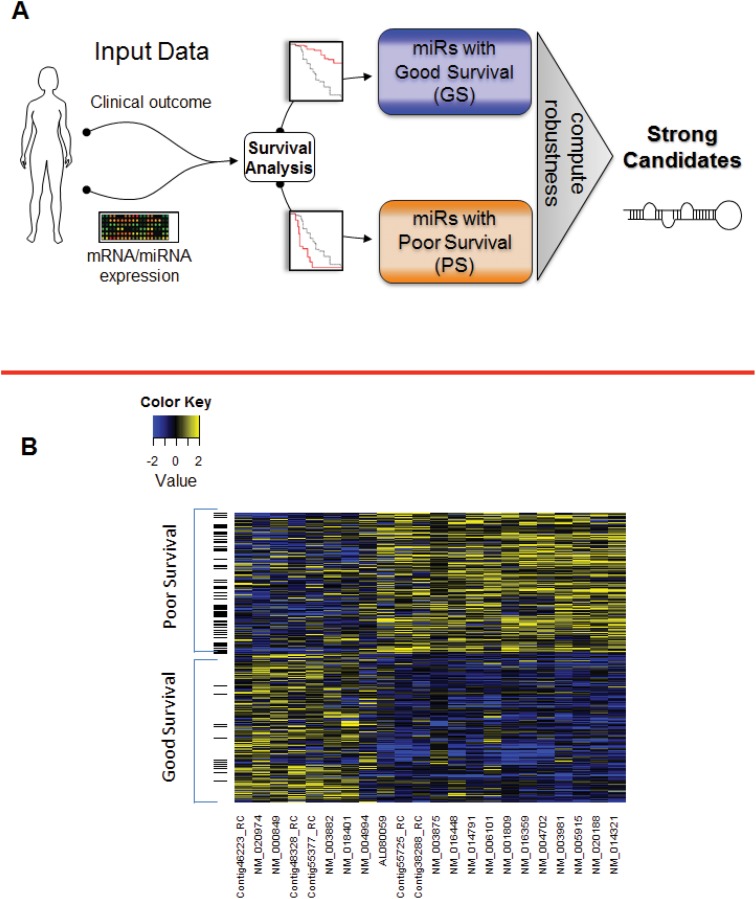
Fig 1. Workflow of our robust selection algorithm (RSA) and validation of the RSA using previously published datasets.
(A) Schematic displaying the overview of the RSA. The inputs are clinical data and miRNA expression data; the outcomes are candidate miRNAs correlated with either good or poor survival. (B) Validation of the RSA using previously published gene signatures correlated with survival outcomes. We applied RSA to breast cancer dataset in Martin et al. And looked at the overlap of genes correlated with good and poor survival computed by RSA and from their results. Heatmap of these overlapping genes was drawn displaying the high gene intensity in yellow and low gene intensity in blue.