Figure 2. Assembly of complete 45S units.
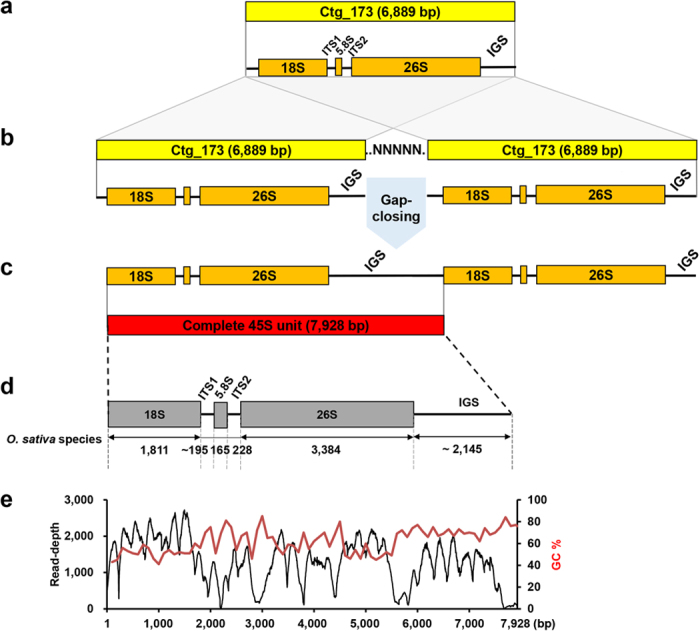
(a–c) Schematic diagram of the method used to obtain a complete 45S unit. (a) A draft single contig included the 45S transcription unit and occasionally part of the IGS. In this example, Ctg_173 assembled using a rice dataset contained a partial IGS. (b) To obtain the full-length IGS, a hypothetical tandem array was constructed using two copies of the contig and intervening Ns. Through a gap-closing process, the Ns were filled in by nucleotide sequences originating from IGS regions. (c) If the IGS remains partial, adjustment of the intervening N length and repeated gap-closing will be necessary. Ultimately, a complete 45S unit with the full-length IGS can be obtained. (d) Structure of the complete 45S unit of Oryza species. (e) Status of read mapping on the assembled 45S units. The Os5 dataset was mapped again to assembled single contigs covering the entire 45S unit sequence (black line). Red line indicates GC content per 100-bp unit length.
