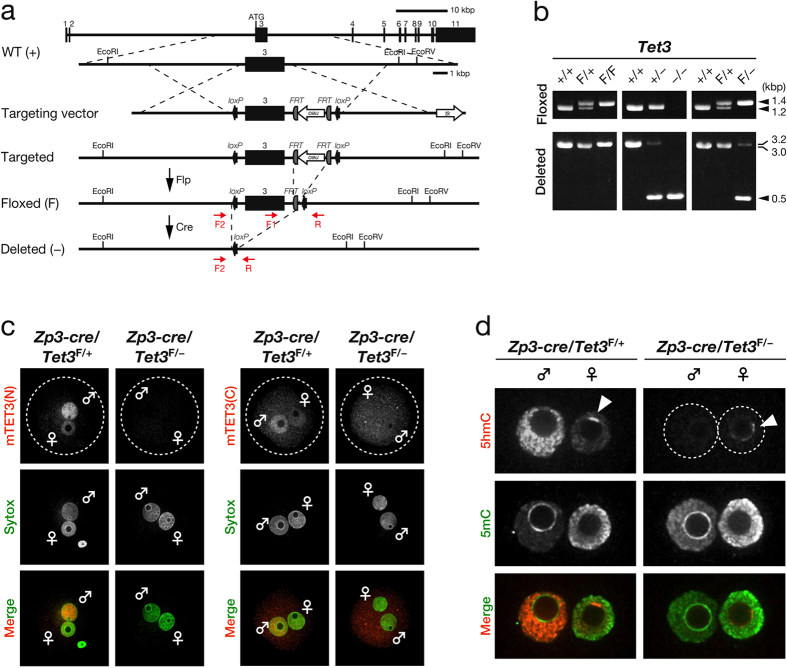
Figure 1. Generation of TET3-depleted oocytes and loss of 5mC oxidation in zygotes derived therefrom.
(a) Strategy for targeted disruption of mouse Tet3. Exons and primers for genotyping are shown as numbered black boxes and red arrows, respectively. See Methods for details. (b) Representative genotyping of mice by PCR analysis. The primers F1 and R were used for detection of WT (1.2 kbp) and floxed (1.4 kbp) alleles, whereas primers F2 and R were used for that of WT (3.0 kbp), floxed (3.2 kbp), and deleted (0.5 kbp) alleles. (c) Representative images showing depletion of maternal TET3 in zygotes derived from oocytes of [Zp3-cre, Tet3F/–] female mice compared with those derived from oocytes of [Zp3-cre, Tet3F/+] female mice. Zygotes were stained with anti-mTET3(N) or anti-mTET3(C) (red in merged images). Pronuclei were revealed by staining of DNA with Sytox (green in merged images). Dashed circles, ♂, and ♀ indicate the rim of the zygote, the paternal pronucleus, and the maternal pronucleus, respectively. (d) Representative images showing the loss of 5hmC in both pronuclei of zygotes derived from oocytes of [Zp3-cre, Tet3F/–] female mice compared with those derived from oocytes of [Zp3-cre, Tet3F/+] female mice. Pronuclei were stained with anti-5hmC (red in merged images) and anti-5mC (green in merged images). ♂, ♀, dashed circles, and arrowheads indicate the paternal pronucleus, the maternal pronucleus, the rim of each pronucleus, and the periphery of the nucleolus-like body, respectively.