Fig. 3.
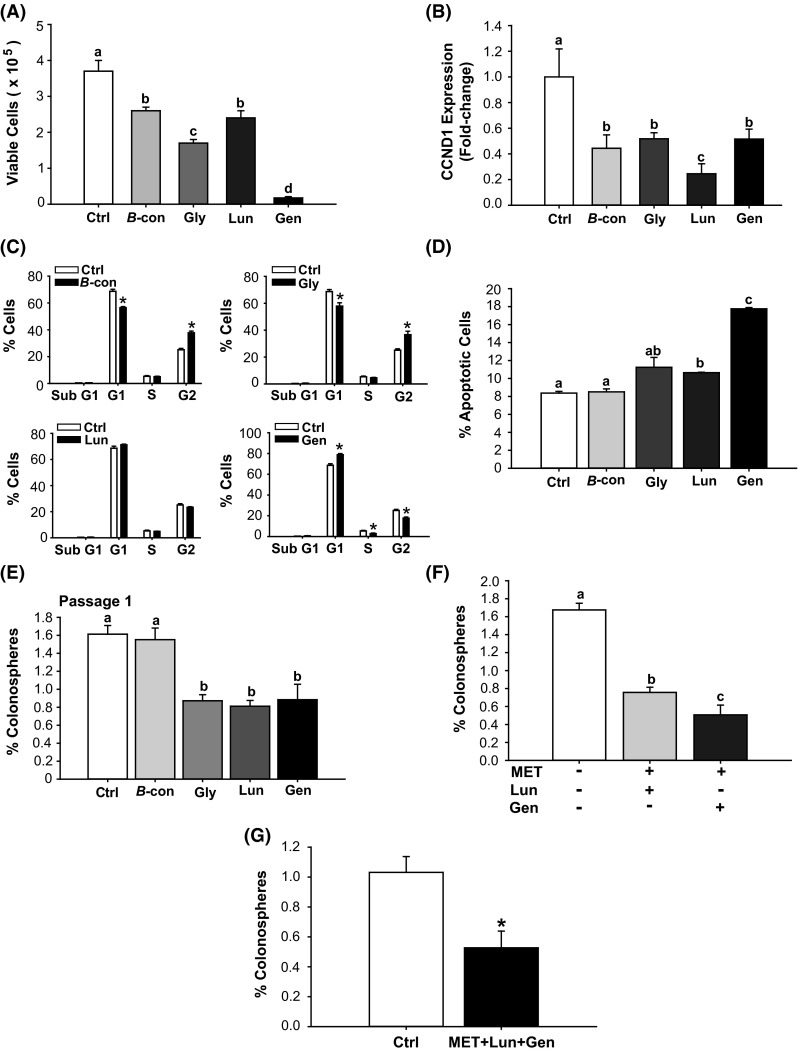
Bioactive soy factors are anti-proliferative, pro-apoptotic and colonosphere inhibitory in HCT116 cells. a HCT 116 cells were treated with the indicated bioactive factors (Gen 2 µM, Lun 2 µM, β-con 3 µM, and Gly 3 µM), and cell viability was measured 48 h later. b Transcript levels of cyclin D1 (CCND1) were determined 48 h after treatment with individual soy factors. Data (mean ± SEM) represent fold-change in normalized expression relative to control; bars with distinct letters differed from control values (P < 0.05). c Cell cycle distribution, determined by fluorescence-activated cell sorting, showed lower percentage of cells in the S and G2 phases and greater percentage of cells in G1, after Gen treatment. β-con and Gly induced lower percentages of cells in G1 and greater percentage of cells in G2. Lun had no effect on cell cycle parameters. Values are means ± SEMs from two independent experiments with each performed in quadruplicate. *P < 0.05 relative to control for each stage of cell cycle. d HCT116 cells were treated with 2 µM (Gen, Lun) or 3 µM (β-con, Gly) of soybean factor and 48 h later, were analyzed for the percentage of apoptotic cells. Lun and Gen induced cell apoptosis (Gen > Lun). Means with distinct letters differed at P < 0.05. e HCT116 cells were treated with soy factors (described above) or vehicle, which were added to plating medium in low attachment plates. Gly, Lun, and Gen diminished the percentage of P1 colonospheres. Means with different letters differed at P < 0.05. Data (mean ± SEM) are from three independent experiments, with each carried out in quadruplicate. f HCT116 cells were treated with MET + Lun or MET + Gen and evaluated for percentage of P1 colonosphere formation. Means with distinct letters differed at P < 0.05, (two independent experiments; each performed in quadruplicate). g Co-addition of MET + Gen + Lun repressed P1 colonosphere formation by 50 %. Data are mean ± SEM, two independent experiments each performed in quadruplicate. *P < 0.05 between groups
