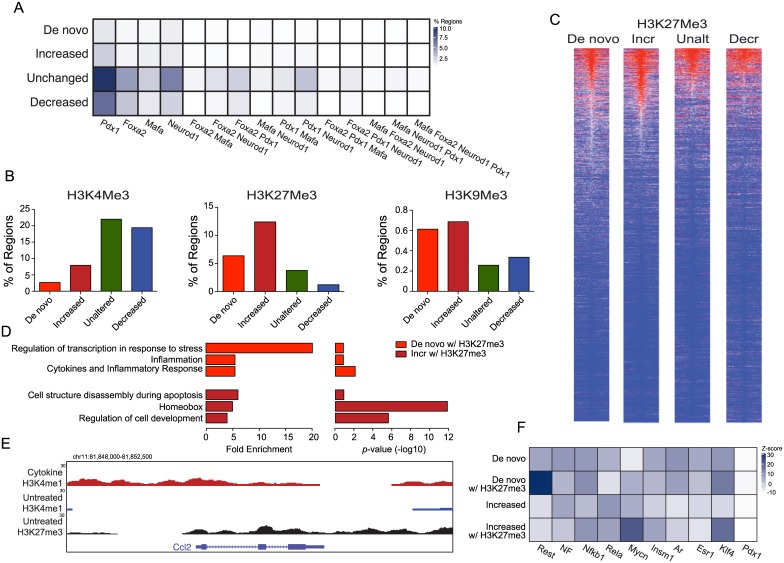
Fig 2. IFNγ, Il-1β, and TNFα-induced de novo and increased loci are frequently H3K27me3 enriched prior to cytokine exposure.
(A) Heatmap of the fraction of de novo, increased, unaltered, and decreased loci bound by the indicated transcription factor combinations. (B) Bar graph of the fraction of de novo, increased, unaltered, and decreased loci pre-marked by H3K4me3 (left graph), H3K27me3 (centre graph), and H3K9me3 (right graph). (C) Heatmaps of H3K27me3 read density in ±10-kb regions centred on the identified de novo, increased (Incr), unaltered (Unalt), and decreased (Decr) loci in untreated islets. H3K27me3 read density is represented by the intensity of blue and red in the heatmaps: dark red indicates high read density while dark blue indicates low read density. (D) Enrichment level (left graph) and p-values (right graph) of gene ontology (GO) terms enriched using genes associated with the identified de novo and increased loci identified as enriched for H3K27me3 in untreated islets. (E) UCSC genome browser view of the genomic region around Ccl2. H3K4me1 enrichment data from IFNγ, Il-1β, and TNFα treated islets is shown in red, from untreated islets in blue, while H3K27me3 data from untreated islets is shown in black. All tracks are set to show a coverage depth range of 0 to 30. (F) Heatmap of the fraction of de novo, increased, unaltered, and decreased loci containing sequences that match the indicated binding motifs.