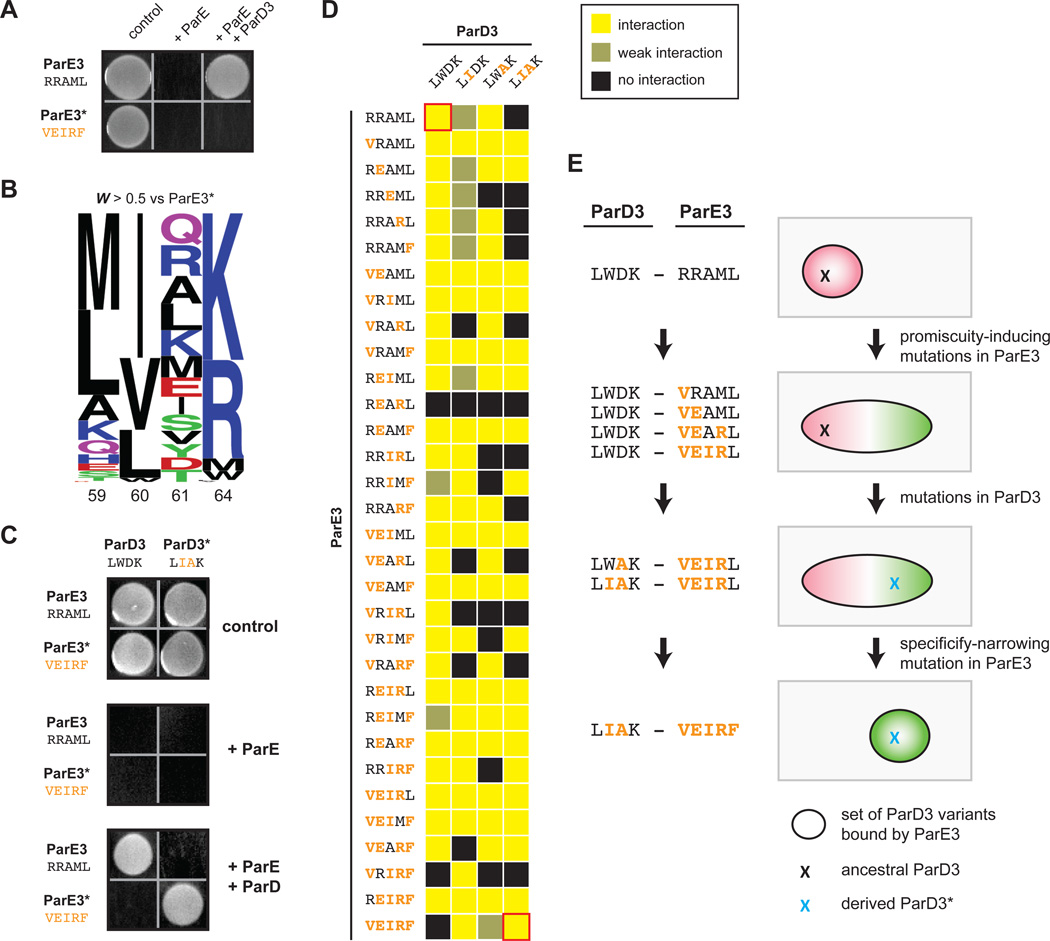
Figure 7. Mutational trajectories to an orthogonal ParD3*-ParE3* pair.
(A) ParE3* is insulated from antitoxin ParD3. A plasmid containing either ParE3 or ParE3* was co-transformed into E. coli with a plasmid expressing ParD3, and cells were plated on medium that induces or represses expression of the toxin and antitoxin.
(B) Frequency logo for ParD3 library variants with high fitness against ParE3* (WE3* > 0.5).
(C) ParE3*-ParD3* is insulated from the wild-type ParD3-ParE3 pair.
(D) Toxicity-rescue interaction assays for all ParD3 and ParE3 mutant combinations. Top left, wild-type ParD3-ParE3 pair; bottom right, orthogonal ParD3*-ParE3* pair. Promiscuous ParE3 intermediates are those capable of interacting with both ParD3 and ParD3*.
(E) Example of a series of single substitutions that lead to the insulated ParE3*-ParD3* system while retaining the toxin-antitoxin interaction at each step by first expanding the specificity of ParE3, followed by changes in ParD3, and finally by restricting the specificity of ParE3.
Also see Fig. S7.