Figure 2. Elements of Pcdh cis and trans binding.
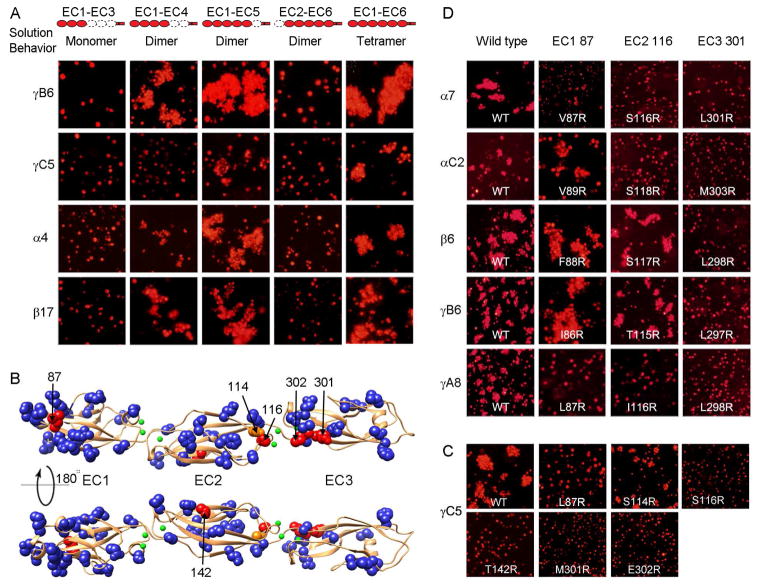
A) Correlating multimerization states of truncated Pcdh proteins with their cell-cell recognition properties. Cells transfected with Pcdh deletion series plasmid constructs were tested for aggregation. With the exception of EC2-EC6 Pcdh fragments and PcdhγC5 EC1-EC4, all deletion proteins that formed oligomers in solution also mediated cell aggregation. Full-length Pcdhα4 include the EC6 domain from PcdhγC3 so it could be delivered to cell surface.
B) Probing homophilic interaction interface by arginine-scanning mutagenesis. Residues mutated to arginine are drawn in space filling representation. In blue are mutations that did not disrupt recognition, in orange are mutations that weakened recognition and in red are mutations that abolished cell-cell recognition. Excluding residue 142, all the effective arginine mutants are located along one side of the molecule.
C) Cell aggregation experiments showing the mutations in part (B) that weakened or abolished interactions. See also Figure S2C.
D) In other Pcdh isoforms, residues analogous to the effective PcdhγC5 arginine mutants had similar effects on the cell-cell recognition in the majority of cases.