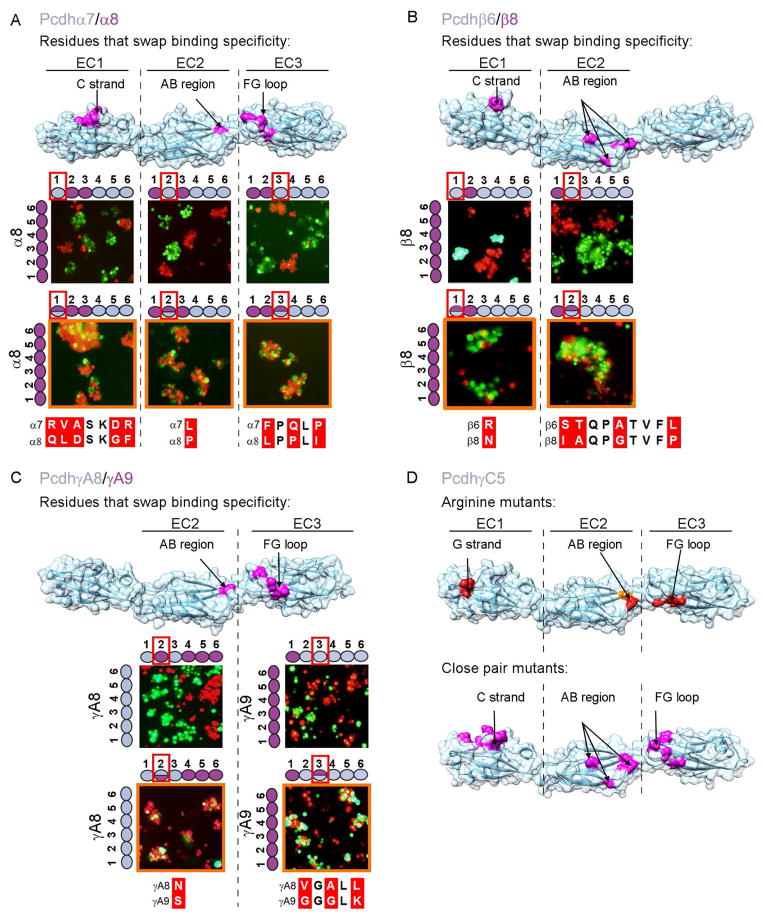
Figure 5. Structural elements of the canonical Pcdh trans binding interface.
A–C) Assessing specificity-determining residues. Binding properties of wild-type isoforms (left side of each panel) or constructs with shuffled residues (top of each panel) were tested separately for each EC domain. Cases in which shuffled residues swapped specificities are indicated by an orange outline. Residues shuffled between closely related isoforms are shown in magenta on surface representations of the Pcdhα7, Pcdhβ6, and PcdhγA8 structures. Sequence alignments of shuffled regions are shown. See also Figure S4.
D) Correspondence between trans interface residues identified by arginine scanning and close-isoform pair analysis. Single arginine mutant residues that abolish or diminish homophilic binding, highlighted in red and orange respectively, are found in the same structural regions as the shuffled residues (see also Figure 2). Residues that swap binding specificity between closely related isoforms are shown in magenta on surface representations of the Pcdh-γC5 crystal structure.