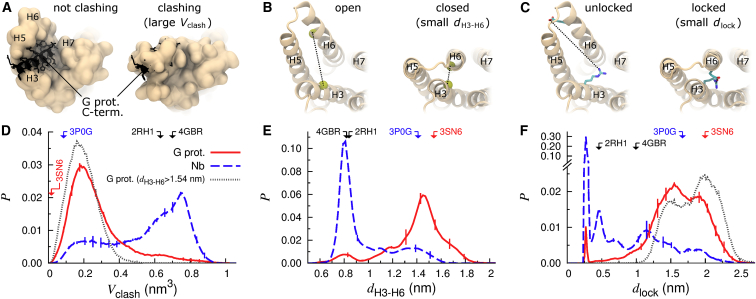
Figure 2.
Receptor deactivation. (A–C) Cytosolic view of representative structures from simulations depicting (left) active and (right) inactive states of the receptor for each of the following deactivation metrics: (A) volumetric clash between the (orange) simulated apo receptor and the (black) C-terminal helix of a Gα protein based on postsimulation modeling, Vclash (see Supporting Materials and Methods); (B) approach of the cytosolic ends of H3 and H6 measured by the distance between (yellow spheres) Cα atoms of R3.50 and L6.34, dH3–H6; and (C) formation of the ionic lock as measured by the minimum Nη-Oε distance between (sticks) R3.50 and E6.30, dlock (also depicted in Movie S1). (D–F) Probability histograms of deactivation metrics in simulations of (solid red line) G-protein- and (dashed blue line) nanobody-derived structures. Dotted gray lines in parts (D) and (F) represent the data from G-protein-derived simulations when dH3–H6 ≥ 1.54 nm, the crystallographic active-state displacement. Vertical bars represent standard error. Arrows and PDB identifiers denote abscissa values for selected crystal structures. To see this figure in color, go online.