Figure 3.
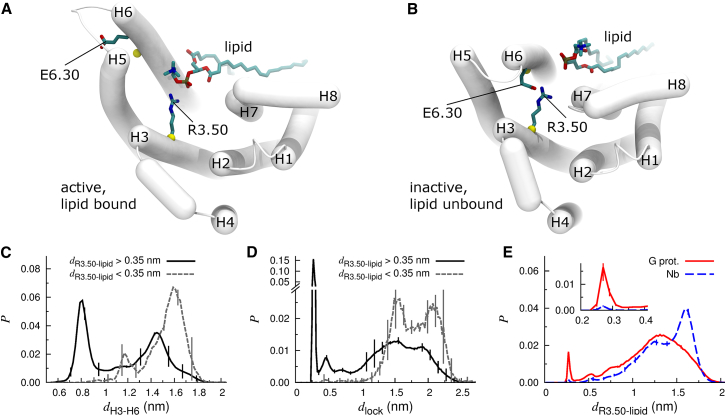
Zwitterionic lipid binding to the R3.50 component of the ionic lock. (A and B) Representative conformations of (A) active, lipid bound and (B) inactive, lipid unbound states. Cα atoms used in evaluation of dH3–H6 are shown as yellow spheres. (C and D) Probability histograms of (C) dH3–H6 and (D) dlock for (solid black line) lipid-unbound and (dashed gray line) lipid-bound states. (E) Probability histograms of the minimum Nη-O distance between R3.50 and a lipid, dR3.50-lipid, for simulations of (solid red line) G-protein- and (dashed blue line) nanobody-derived structures. Probability density maps depicting dR3.50-lipid vs. dH3–H6 and dlock are presented in Fig. S4. To see this figure in color, go online.