Fig. 1.
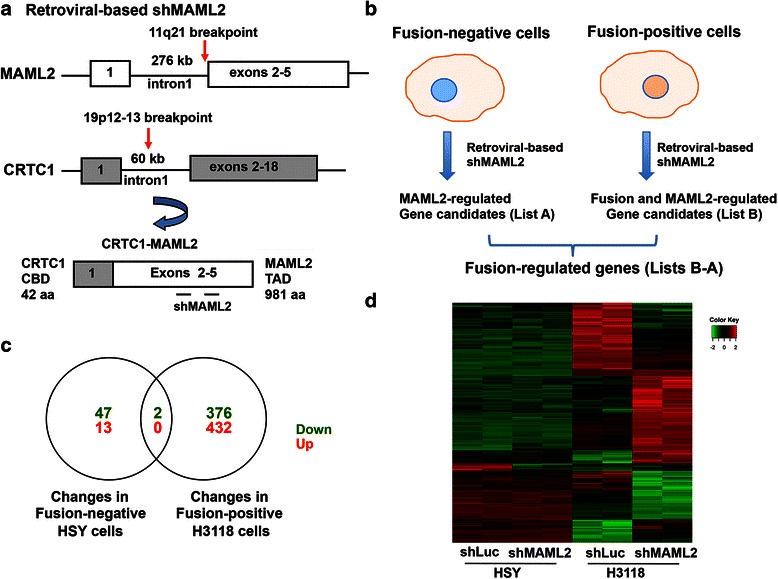
Transcriptional profiling analysis revealed target gene candidates regulated by the CRTC1-MAML2 fusion oncogene in human MEC cells. a The t(11;19) translocation fuses exon 1 of the CRTC1 gene to exons 2–5 of the MAML2 gene and generates the CRTC1-MAML2 fusion consisting of CRTC1 CREB binding domain (CBD) and MAML2 transcriptional activation domain (TAD). b The strategy in identifying CRTC1-MAML2-regulated genes by microarray analysis was shown. Fusion-negative cells (HSY) and fusion-positive cells (H3118) were infected with retroviral-mediated MAML2 shRNA-IRES-GFP (co-expressing MAML2 shRNA and GFP) or luc shRNA-IRES-GFP (co-expressing luciferase shRNA and GFP that serves as a control) for 72 h. GFP-positive cells were then FACS-sorted, and RNA samples were collected for microarray analysis using HG-U133 Plus 2.0 arrays. Comparison of MAML2-knockdown HSY cells and the control cells resulted in MAML2-regulated gene candidates (list A), while comparison of fusion/MAML2-knockdown H3118 cells and the controls led to fusion/MAML2-regulated genes (list B). Based on these two lists of target genes, fusion-regulated genes were identified by filtering out the list A from the list B (List: B-A). c, d Venn diagram (c) and heatmap (d) show differentially expressed genes in both fusion-negative HSY and fusion-positive cells after shMAML2 transduction. Two biological replicates were analyzed. This analysis led to the identification of a total of 808 fusion-regulated candidate genes, with 376 down-regulated and 432 up-regulated genes
