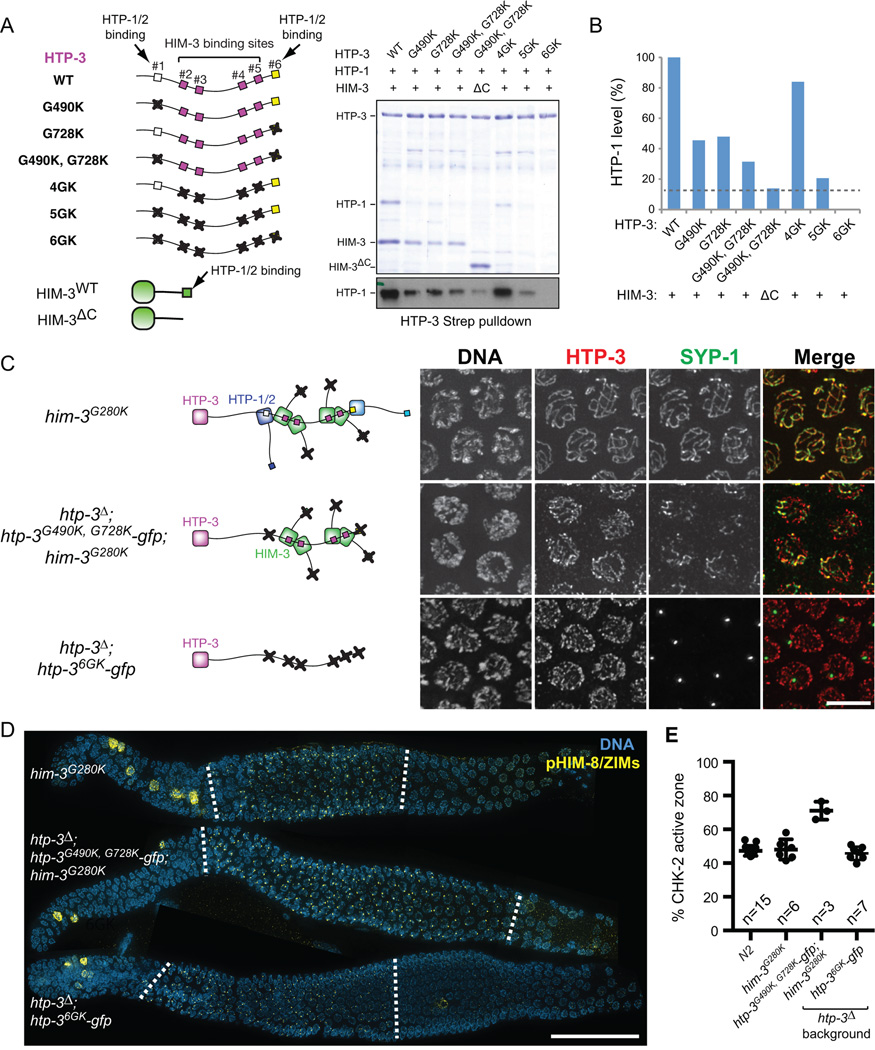
Figure 6. Axis localization of HTP-1/2 and HIM-3 is essential for feedback control of CHK-2.
(A) Strep-tagged HTP-3, either wild-type or GK mutants, was coexpressed in E. coli with untagged HTP-1 and wild-type or C-terminally truncated HIM-3 (ΔC; aa 1–245), and purified using Strep-resin to isolate proteins associated with HTP-3. Coomassie staining of SDS-PAGE shows the HTP-3-associated proteins. A Western blot for HTP-1 is shown below. (B) The amount of HTP-1 bound by HTP-3 was quantified based on the HTP-1 Western blot. (C) Schematics of HORMA domain protein assembly in the indicated genotypes are shown in the left. Mid-pachytene nuclei were stained for DNA (white), HTP-3 (red), and SYP-1 (green). Scale bar, 5 µm. (D) Immunofluorescence images of gonads dissected from him-3G280K, htp-3Δ; htp-3G490K, G728K-gfp; him-3G280K, and htp-3Δ; htp-36GK-gfp showing DNA (blue) and pHIM-8/ZIMs (yellow). White dotted lines indicate the CHK-2 active zone. Scale bar, 50 µm. (E) Quantification of the CHK-2 active zone in indicated strains. Numbers of gonads scored are indicated below. htp-3Δ; htp-3G490K, G728K-gfp; him-3G280K mutants show significant differences from N2 (p<0.0001), while others do not (him-3G280K, p=0.9608; htp-3Δ; htp-36GK-gfp, p=0.7806), analyzed by ordinary one-way ANOVA.