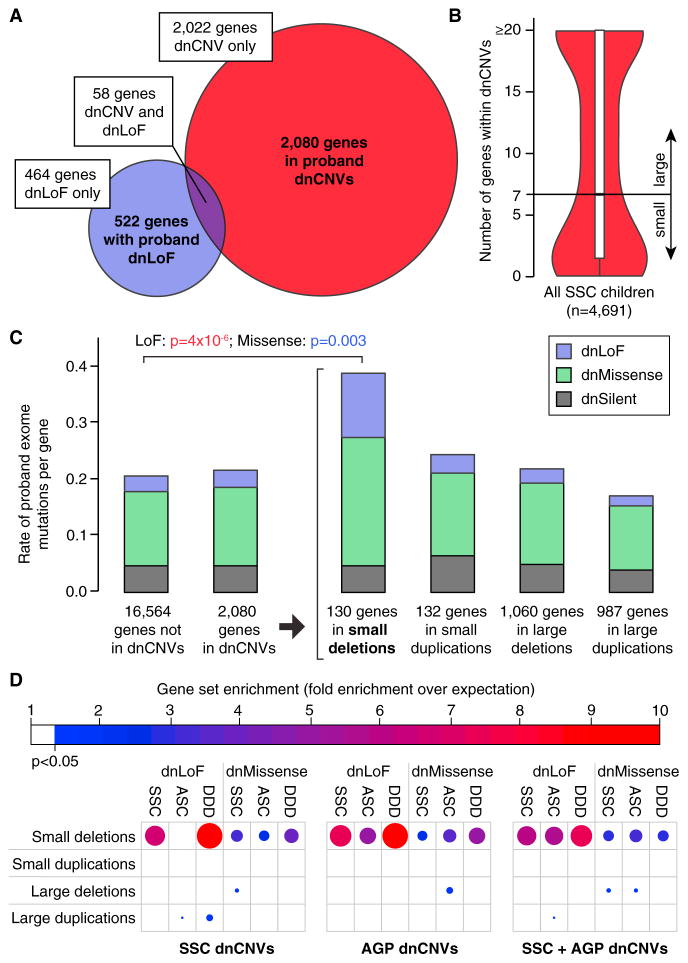
Figure 5. Small De Novo Deletions Are Enriched for Exome Mutations.
(A) 2,080 unique genes are identified within pro-band dnCNVs (red) and 522 unique genes have dnLoFs in probands (purple); 58 unique genes are observed in both these datasets.
(B) The median number of genes within validated dnCNVs in the SSC is seven; this threshold is used to distinguish small and large dnCNVs.
(C) The number of de novo mutations per gene observed with exome sequencing of the SSC and ASC are shown in different groups of genes based on dnCNV overlap. Mutation rates are normalized for gene mutability based on gene size and GC content. Exome mutations are divided into silent (gray), missense (green), and LoF (purple). No excess of exome mutations is observed in the 2,080 genes within dnCNV regions compared to the 16,564 genes outside of dnCNVs. Dividing the dnCNV regions by size (≤7 genes versus >7 genes) and type (deletion versus duplication) reveals strong enrichment for dnLoF (p = 4 × 10−6, Fisher Exact Test) and dnMissense (p = 0.003) in small de novo deletions only.
(D) The enrichment of genes within dnCNVs is shown by the size and shade of the circle (red and large = high degree of enrichment; blue and small = modest degree of enrichment); only results reaching nominal significance (hypergeometric test) are shown. Small de novo deletions show consistent enrichment for dnLoF and dnMissense mutations across three cohorts: SSC, Autism Sequencing Consortium (ASC), and Deciphering Developmental Disorders (DDD). This result is observed for dnCNVs detected in the SSC and Autism Genome Project (AGP) independently and in combination.