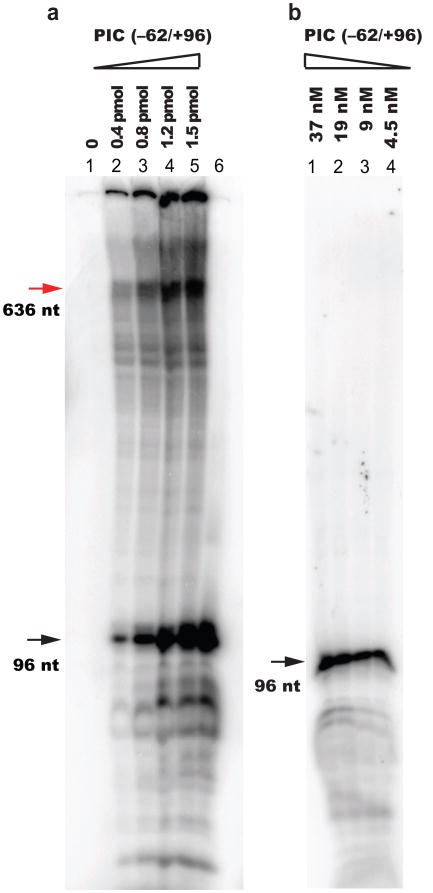
Extended Figure 3. Run-off transcription under single-molecule assay conditions.
(a) 0.1 pmol of the isolated PIC formed on the SNR20* fragment (−62/+636) fused to the 2.7kbp DNA handle was transcribed in the presence of increasing amount of PICs assembled on the same promoter SNR20* (−62/+96) without handle. The SNR20* fragment fused to the DNA handle failed to support transcription (lane 1), but the transcription activity was restored (red arrow) when PICs without handle were supplemented into the mix (lanes 2–5). Run-off transcription from the SNR20* promoter without handle is indicated (black arrow). We adopted a 25:1 ratio of PICs assembled on SNR20* without handles to PICs assembled on SNR20* with handles for the single-molecule study. (b) 1.5 pmol of PIC formed on the SNR20* fragment (−62/+96) was transcribed at the concentrations indicated above each lane. As the concentration of PIC was reduced from 37 nM to 4.5 nM, the transcription efficiency (run-off band, black) decreased from ∼18% to 2-3%. The extremely low concentrations employed in single-molecule assays (<1 nM) could not be directly examined on such gels, but we expect that transcription efficiencies remained correspondingly low.