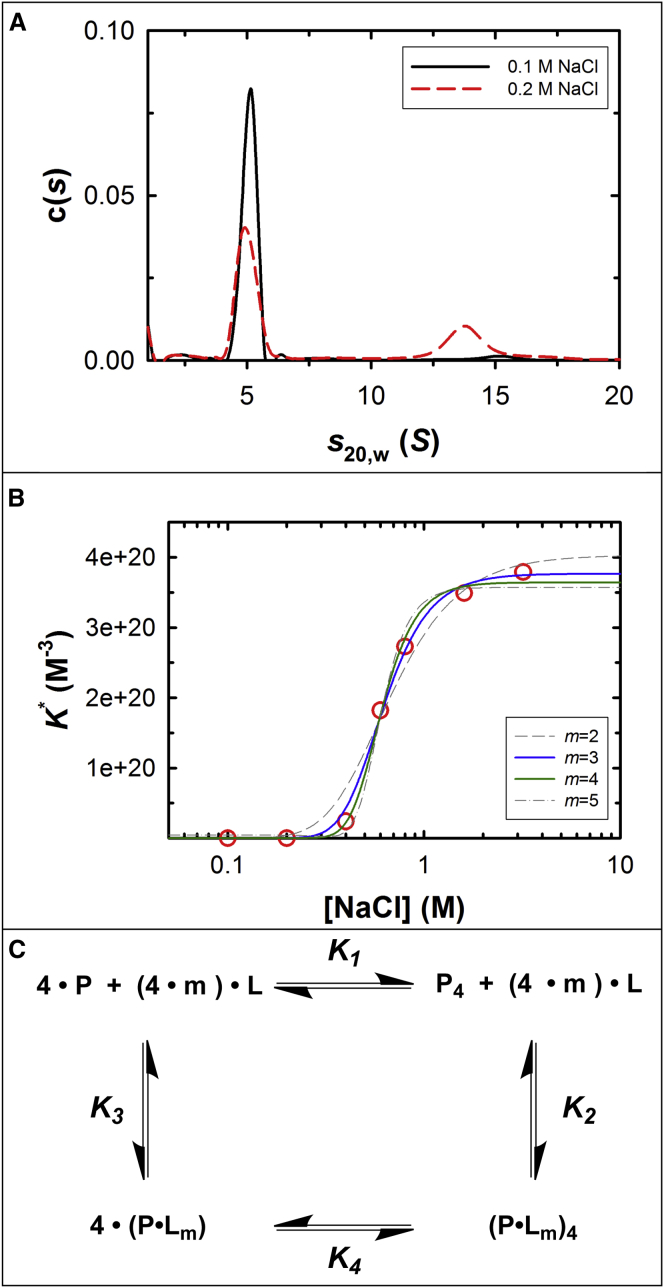
Figure 5.
Dependence of self-association on NaCl concentration. (A) Sedimentation velocity of terminase analyzed by c(s) analysis, showing different species distribution under different NaCl conditions (0.1 and 0.2 M, black solid and red dashed lines, respectively). Note that there is the slight decrease in s20,w of the large species at the higher NaCl concentration, a phenomenon that we have observed previously (30). We interpret this to indicate that the conformation of the tetramer is affected by salt to afford a more extended shape, and thus a smaller s20,w value. (B) (Red circles) Sedimentation equilibrium data presented in Table 1; (superimposed lines) nonlinear least-squares fits to the model presented in (C) using Eq. 4, with m fixed at the indicated value. The results of the NLLS fits are presented in Table 2. (C) Model for thermodynamic linkage of the monomer-tetramer equilibrium and NaCl binding. P is protein monomer, P4 is tetramer, and m is the net number of NaCl (L) binding or releasing in the equilibria. The individual association constants are defined as follows: K1 is the association constant of protomer-tetramer equilibrium with no net NaCl participating; K2 is the association constant of NaCl binding to tetramer; K3 is the association constant of NaCl binding to monomer; and K4 is the association constant of monomer-tetramer equilibrium under saturated NaCl binding condition. The model assumes that the number of ions that bind to the protomer in isolation and in the context of the tetramer is identical. To see this figure in color, go online.