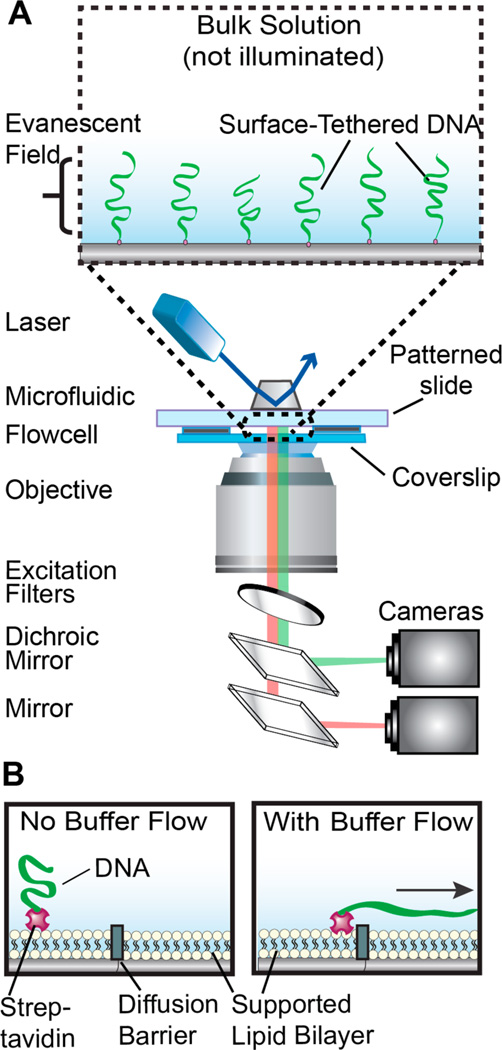
Figure 1.
An illustration of the DNA curtains platform. (A) DNA molecules are immobilized on the passivated surface of a microfluidic flowcell. The DNA is illuminated via a laser beam (488 nm) that impinges on a prism in total internal reflection fluorescence (TIRF) mode, thereby generating an evanescent excitation wave at the interface between the lithographic patterned surface and the imaging buffer. The evanescent wave penetrates ~200 nm away from the micropatterned flowcell surface to selectively illuminate surface-bound DNA and protein molecules. The resulting fluorescent signals propagate through a coverslip and are collected via a high numerical aperture objective, passed through two excitation clean-up filters (490 and 500 long pass; Chroma), and dispersed through a dichromic mirror onto two different charge coupled device (CCD; ANDOR) cameras. (B) Side view of a DNA molecule (green) that is affixed to a lipid bilayer (circles) via a biotin–streptavidin (magenta) linkage. In the presence of buffer flow, the DNA molecule moves within the fluid lipid bilayer and is captured at a Cr diffusion barrier (gray).