Fig 4. Comparing ubiquitin ensembles from simulations and experiments.
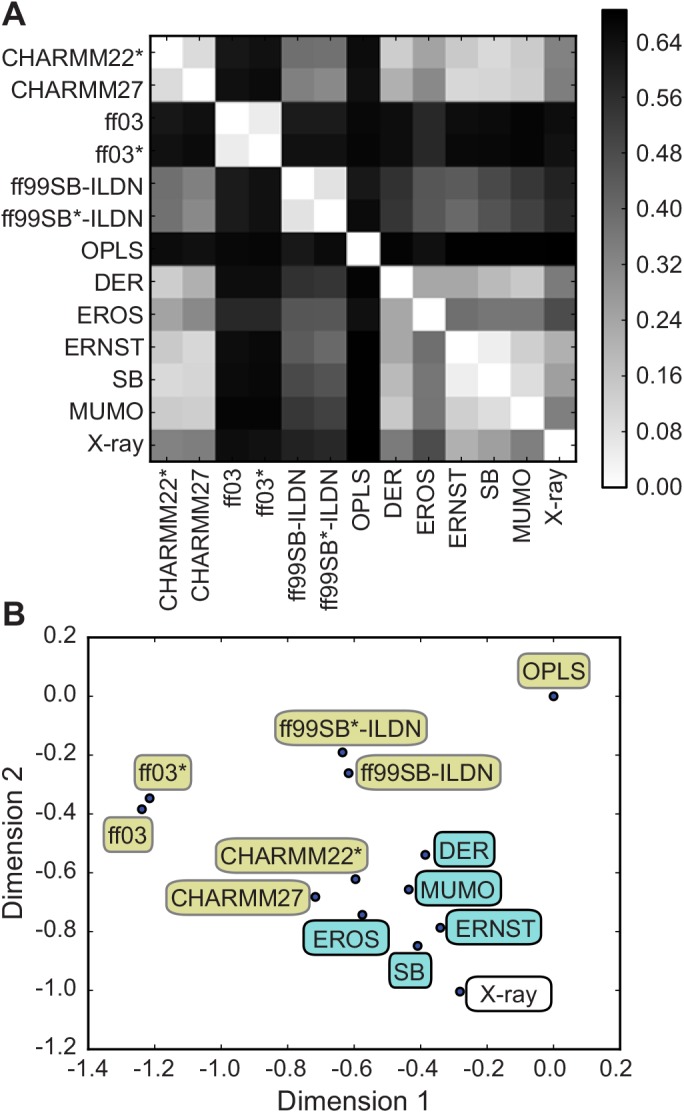
We used ENCORE to compare 13 previously determined conformational ensembles of human ubiquitin: seven ensembles were obtained by molecular dynamics simulations with different force fields, five ensembles were generated via replica-averaged simulations that used experimental NMR data as restraints in molecular simulations and a single ensemble was obtained as a collection of 46 different crystal structures of ubiquitin. (A) Using CES we calculated the pairwise similarity of all 13 ensembles and (B) projected the results into two dimensions. Note how the molecular simulations (yellow labels) result in a broader range of conformational ensembles whereas the ensembles restrained via different kinds of experimental NMR data (blue labels) are all more similar to one another. This observation is evidence of the fact that experimental restraints, when used in replica-averaged simulations, can be thought of as system specific correction to the energy function used, which guides the simulations towards the correct conformational ensemble. Finally, note how the NMR-restrained simulations are also relatively similar to a collection of ubiquitin X-ray structures. This observation reiterates the notion that ubiquitin in solution samples a conformational ensemble that is similar to the variability observed in different ubiquitin structures, and also that such ensembles can be derived relatively robustly by combining NMR data and molecular simulations. Importantly, the five NMR ensembles were obtained using different procedures, force fields and sources and types of experimental data.
