Fig 2. Comparison of the accuracy of mutant interface profile scores formed from different structural alignment methods in predicting ΔΔG of complex formation.
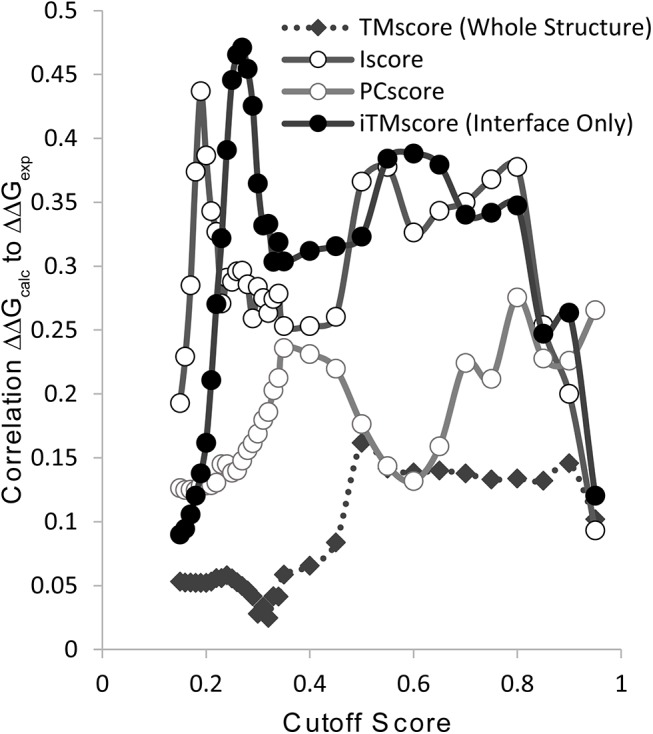
The iTM-score considers only structural similarity at the interface, Iscore considers structural similarity at the interface and the fraction of native contacts preserved, and PCscore considers both physicochemical and structural similarity at the interface. TM-score considers only structural alignment of the mutated monomeric protein. Profiles are constructed from sequences meeting each cutoff and the predicted ΔΔG values are calculated according to Eq 2.
