Fig 3. Dependency of the accuracy of ΔΔG prediction on the number of sequences that can be aligned at the site of the mutation and the formation of an adaptive profile mixing sequences from high and low interface similarities.
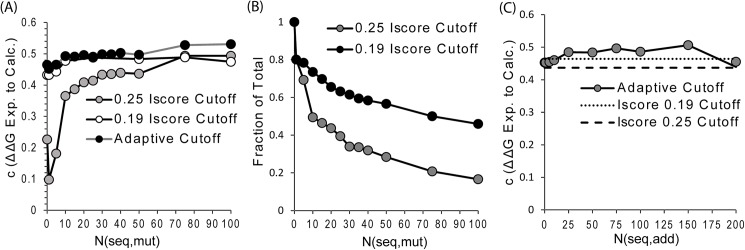
Only single site mutations are considered (81% of the total number of mutations). N seq,mut and N seq, add are the number of sequences that can be aligned at the site of the mutation and the number of lower similarity sequences added to the profile, respectively. (A) Pearson’s correlation c between predicted and experimental ΔΔG values as a function of the number of sequences that can be aligned at the site of the mutation. (B) Fraction of the total number of single site mutations as a function of the number of sequences that can be aligned at the site of the mutation. (C) Improvement in accuracy of an adaptive profile mixing sequences from high and low interface similarities over profiles formed purely using high and low interface similarity cutoffs.
