Fig 11. Accuracy of ΔΔG prediction on a per protein basis after leave-one-protein-out cross-validation for the 24 proteins with more than 10 mutants available based on the standard error of prediction.
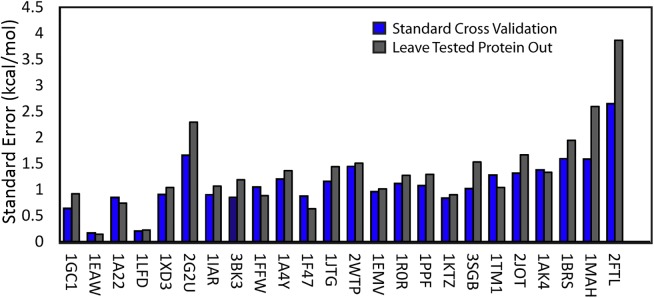
Proteins are arranged left to right in order from the low to high mean experimental ΔΔG value. The mean standard error across the set increases from 1.11 kcal/mol to 1.33 kcal/mol if the tested protein is left out during training.
