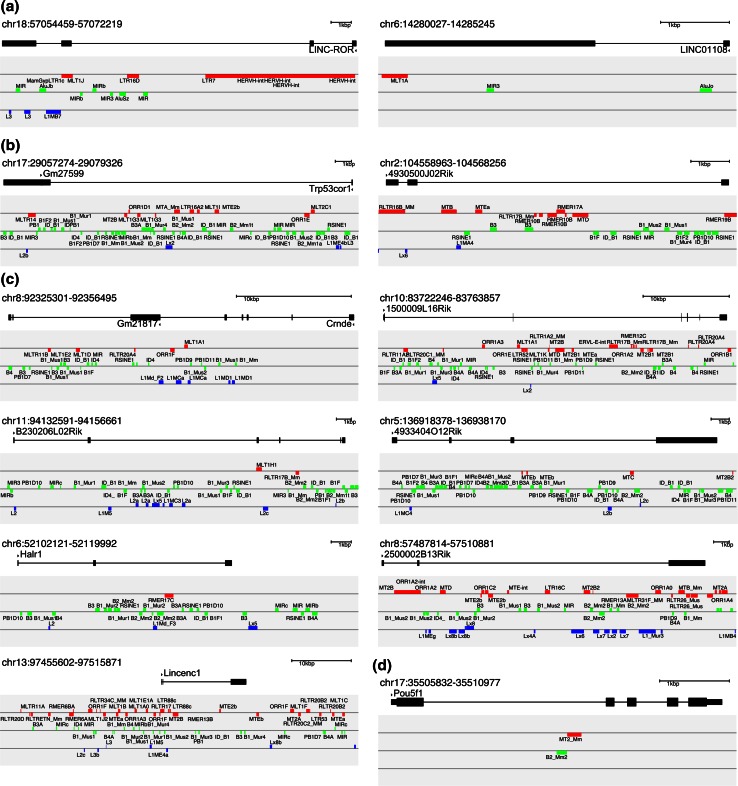
Fig. 4.
Genomic views of selected lncRNAs with demonstrated involvement in reprogramming or ESC maintenance reveal extensive presence of TEs. Genomic views indicate the gene; thick black parts indicate exons, which are connected with thin lines (introns). TEs are indicated across three lines in the gray panel, the top (red) indicates LTR endogenous retroviruses, the middle (green) indicates SINEs, and the bottom (blue) indicates LINEs. TEs above the light gray line are on the positive DNA strand and TEs below the light gray line are on the negative DNA strand. Some duplicate TE labels were removed for clarity. a LncRNAs involved in human iPSC reprogramming, LINC-ROR [89], and LINC01108 (linc-ES3) [91]. b Two mouse lncRNAs involved in reprogramming: Trp53cor1 (lincRNA-p21) [92] and 4930500J02Rik (Ladr83) [95]. c A selected series of lncRNAs involved in the maintenance of mouse ESCs [96]: Cnrde/Gm21817 (Linc1399), 1500009L16Rik (Linc1435), B230206L02Rik (Linc1448), 4933404O12Rik (Linc1543), Halr1 (Linc1547), 2500002B13Rik (Linc1577), and Lincenc1 (Linc1283). d The critical pluripotency gene Pou5f1 is shown for comparison. The protein-coding sequence is indicated with a thicker black line within the exons. Mouse genomic coordinates are mm10 and human are hg38 assemblies