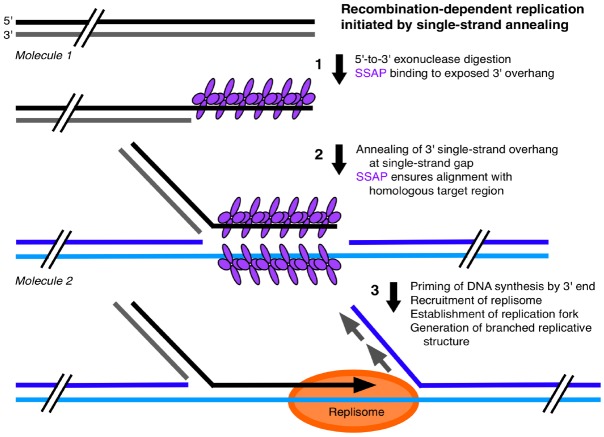
FIGURE 3.
Single-strand annealing mechanism for plastid DNA replication. This single-strand annealing (SSA), recombination-dependent replication model for ptDNA is based on a replication mechanism for herpes virus DNA (Weller and Sawitzke, 2014). (1) A 3′-overhang is generated by 5′-to-3′ exonuclease digestion at the end of a unit-genome-sized monomer. A single-strand annealing protein (SSAP) binds to a 3′-overhang. (2) Annealing of the 3′-overhang of Molecule 1 to a homologous single-strand gap in another ptDNA molecule (Molecule 2). (3) Replication is initiated by priming at the 3′-end, assembly of the replisome, and formation of a replication fork, leading to a branched-linear structure. A similar model with the same or analogous proteins applies to the replication of mtDNA in plants. We propose that replication occurs only with ptDNA attached to the nucleoid-on-membrane using single-strand end-binding proteins. Although we propose that Whirly proteins serve attachment and SSAP functions, other single-strand-binding proteins, such as the OSB and RecA families, may also participate in ptDNA replication. Other replication and recombination mechanisms have been described (Cox, 2007; Marechal and Brisson, 2010; Weller and Sawitzke, 2014; Morrical, 2015).