Figure 3.
iC9-iPSCs Can Efficiently Differentiate into Virus-Specific CTLs
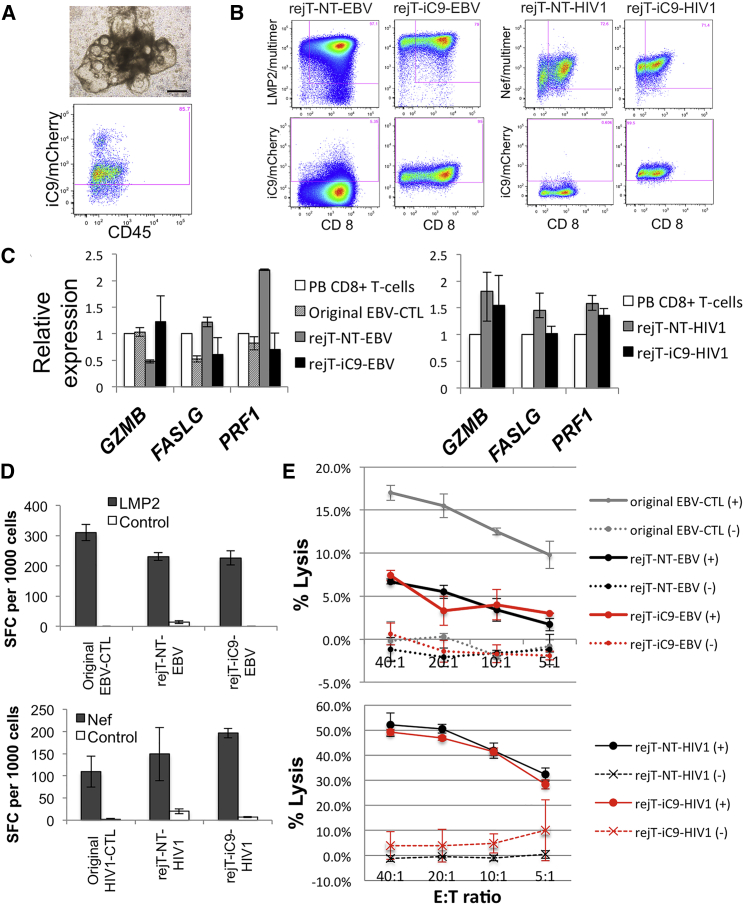
(A) iC9-HIV1-iPSC sacs on day 14 of culture on C3H10T1/2 feeder cells (upper). The scale bar represents 500 μm. One day after sac extraction, extracted hematopoietic progenitor cells were analyzed by flow cytometry. Flow cytometric analysis of iC9/mCherry expression by CD235a−, CD34+, and CD43+ gated cells (lower).
(B) Flow cytometric analysis of peptide-HLA multimer labeling/CD8 expression by iPSC-derived CTLs 14 days after the third stimulation. Expression of iC9/mCherry by these CTLs is shown. The plots are representative of at least five independent differentiation experiments.
(C) Quantitative PCR analysis to compare the expression of cell-lysis molecules in PB CD8+ T cells, original EBV-CTLs, rejT-NT-EBV, and rejT-iC9-EBV (left). The same analysis was also carried out in PB CD8+ T cells, rejT-NT-HIV1, and rejT-iC9-HIV1 (right). Individual PCR results were normalized against 18S rRNA. Data are presented as the mean of three independent experiments ± SD.
(D) IFN-γ production by original EBV-CTLs, rejT-NT-EBV, and rejT-iC9-EBV in the presence of LMP2 peptide was measured using ELISPOT. IFN-γ production by original HIV1-CTLs, rejT-NT-HIV1, and rejT-iC9-HIV1 in the presence of Nef peptide was measured similarly. Data are presented as the mean ± SD and are representative of three independent triplicate experiments.
(E) In vitro 51Cr-release assay of original EBV-CTLs, rejT-NT-EBV, and rejT-iC9-EBV (effectors) and EBV-transformed B lymphoblastoid cell lines (targets) (upper) and that of rejT-NT-HIV1 and rejT-iC9-HIV1 (effectors) and Nef-presenting LCLs (targets) (lower). Data are presented as the mean ± SD and are representative of at least three independent triplicate experiments.
See also Figure S2.