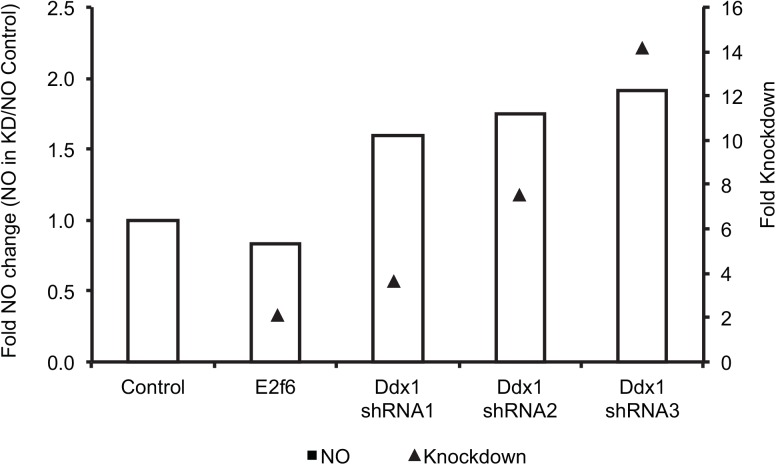
Fig 3. shRNA- mediated Ddx1 knockdown in C57BL/B6J immortalized macrophages relieves NO inhibition.
Fold knockdown of Ddx1 and fold change in NO levels are relative to the Ddx1 expression and the amount of NO in cells transduced with control shRNA (LacZ), respectively. Knocking down E2f6, the other candidate gene at this locus, did not affect the amount of NO produced. Shown are values of NO (μM) fold change obtained from two independent experiments using three different shRNA constructs. The knockdown level is indicated by the black triangles. Fold knockdown was calculated using the 2deltadelta Ct method. The shRNA transductions and NO measurements were done in 3 independent replicates.