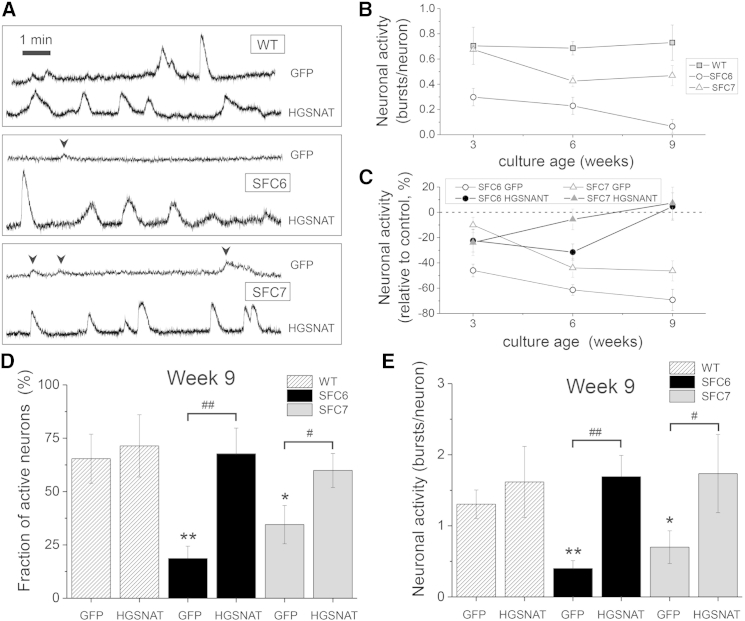
Figure 5.
Altered Neuronal Activity in SFC-Derived Neuronal Networks
(A) Representative traces of spontaneous neuronal activity at week 9 of differentiation analyzed by calcium fluorescence imaging. Top traces correspond to control (GFP-transduced) neuronal networks; bottom traces correspond to gene complemented (HGSNAT-transduced) networks. Sharp increases in the fluorescence signal denote bursting episodes. Firings events of low amplitude (arrowheads) correspond to single spikes.
(B) Neuronal activity (total number of firings per neuron monitored along 30 min recording) of three independent untransduced cultures, at 3, 6, and 9 weeks after differentiation for each cell line and at each time point.
(C) Relative change in activity of control (GFP-transduced) or gene complemented (HGSNAT-transduced) SFC6 and SFC7 neuronal networks with respect to their equivalent WT networks. Three independent cultures were analyzed for each cell line at each time point.
(D) Fraction of active neurons at week 9 in control (GFP-transduced) or gene complemented (HGSNAT-transduced) WT, SFC6, and SFC7 neuronal networks. Three independent cultures were analyzed for each cell line at each time point.
(E) Neuronal activity of control (GFP-transduced) or gene complemented (HGSNAT-transduced) WT, SFC6, or SFC7 neuronal networks at week 9 of differentiation. In (B) through (E), error bars are root-mean-square error. Three independent cultures were analyzed for each line at each time point. ∗p < 0.05 (patients versus WT), ∗∗p < 0.01 (patients versus WT), #p < 0.05 (GFP- versus HGSNAT-transduced), ##p < 0.01 (GFP- versus HGSNAT-transduced).