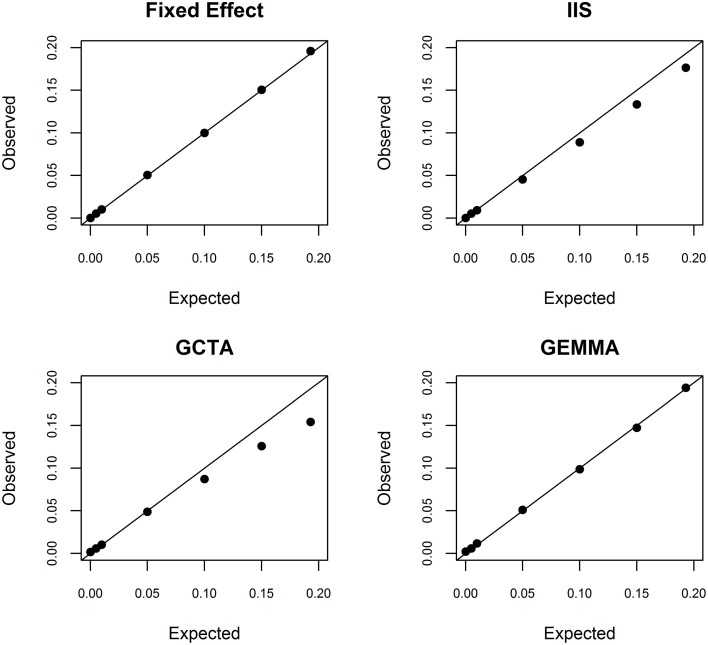
Figure 2.
Simulation study of bias in the random effects model. Conditional on local ancestry at rs2814778, we simulated a continuous phenotype with a known proportion of phenotypic variance explained by a single causal locus and the remainder of the phenotypic variance being random noise. We randomly generated 100 independent data sets. We then used the fixed effects model (top left), ancestral similarity defined by identity in state (top right), centered and scaled ancestral similarity as defined by GCTA (bottom left), and centered ancestral similarity as defined by GEMMA (bottom right) to estimate the proportion of phenotypic variance explained by local ancestry using similarity estimated genome-wide.