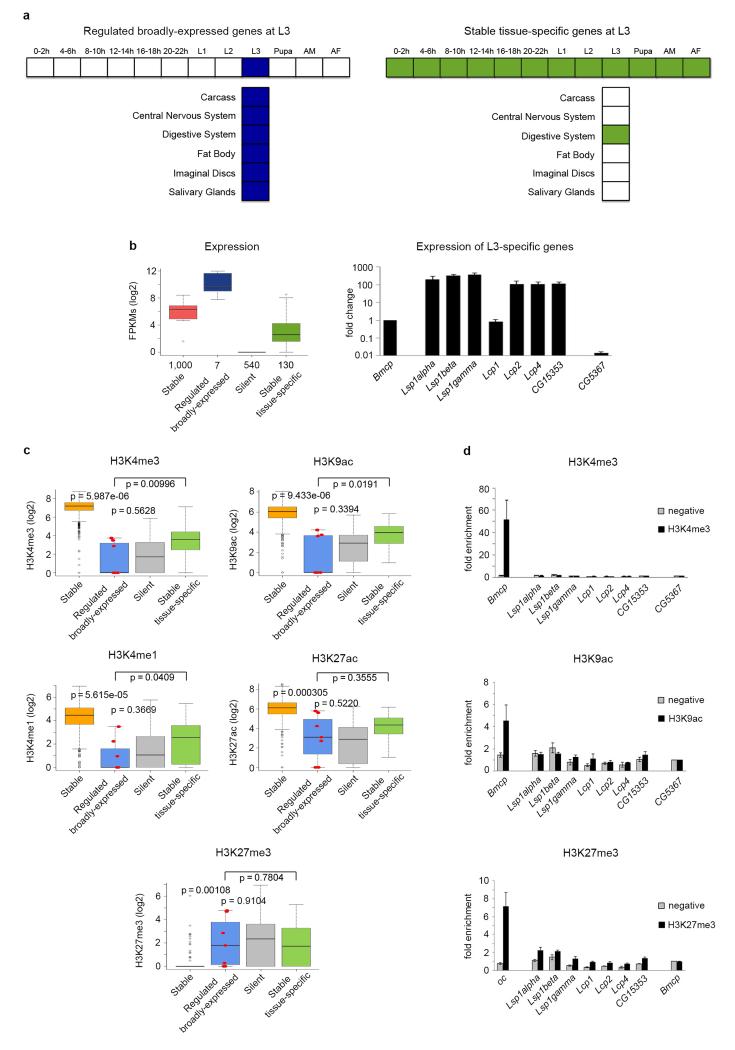
Figure 2. Gene expression and histone modifications in regulated broadly-expressed and stable tissue-specific genes at third instar larvae.
a, Diagrams of developmentally regulated genes broadly-expressed across multiple tissues at third instar-larvae L3 (left panel), and stable genes expressed in only one tissue at L3 (right panel). b, Gene expression levels at L3 measured by whole organism RNASeq (left panel). The number of genes in each category is given under the boxplots. The bottom and top of the boxes are the first and third quartiles, and the line within, the median. The whiskers denote the interval within 1.5 times the IQR from the median. Outliers are plotted as dots. Validation by qPCR of the expression at L3 of regulated broadly-expressed genes compared to a stable gene (Bmcp) and a silent gene (CG5367) (right panel). Error bars represent the Standard Error of the Mean (SEM) from three independent replicates. c, Levels of H3K4me3, H3K9ac, H3K4me1 and H3K27ac on whole L3 individuals. The seven regulated genes broadly-expressed at L3 are depicted as red dots within the boxplots. P-values were computed using the Wilcoxon test (two-sided). d, Validation by individual ChIPs and qPCR of H3K4me3 and H3K9ac in regulated genes broadly-expressed at L3. H3K4me3 and H3K9ac ChIPs are represented as enrichment of the marks over the silent gene (CG5367). Error bars represent the SEM from three independent replicates.