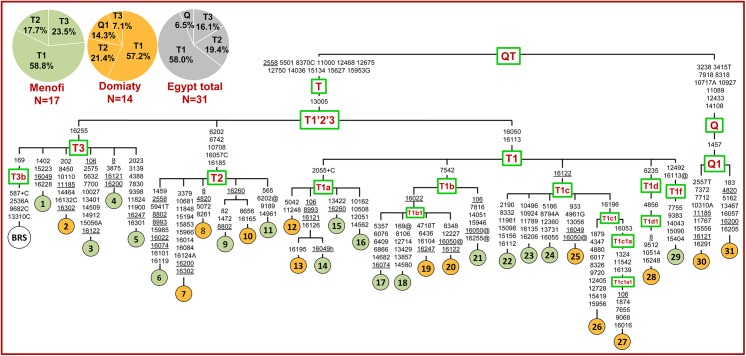
Fig 1. Tree of mitogenomes from Egyptian cattle.
Sequences #1–19, #25, #30–31 have been determined in this study, while sequences #20–24 and #26–29 were previously reported [27]. GenBank accession numbers are reported in S1 Table. This tree was built as previously described [22–24, 27]. The hypervariable insertion of a G at np 364, the length variations in the C tract scored at np 221 and the A tract scored at np 1600 were not used for the phylogeny construction. The position of the Bovine Reference Sequence (BRS) [36] is indicated for reading off-sequence motifs. Branches display mutations with numbers according to the BRS; they are transitions unless a base is explicitly indicated for transversions (to A, G, C, or T) or a suffix for indels (+, d) and heteroplasmy (h). Recurrent mutations within the phylogeny are underlined and back mutations are marked with the suffix @. Note that the reconstruction of recurrent mutations in the control region is ambiguous in a number of cases. The pie charts summarize haplogroup frequencies in the Menofi (green) and Domiaty (orange) breeds.