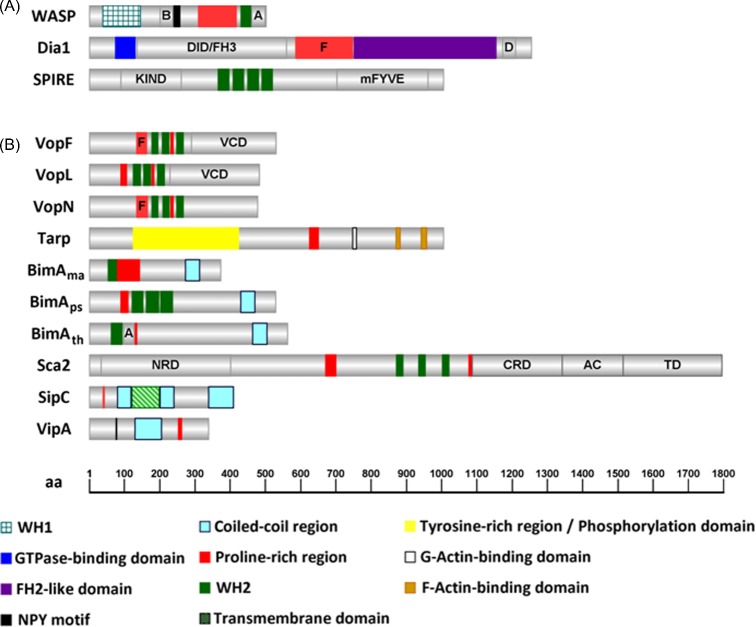
Figure 1.
Graphical representation of the domain organization of proteins involved in actin nucleation. (A) Eukaryotic actin nucleators: WASP, Dia1 and SPIRE (Homo sapiens; Sarkar et al. 2014; Goode and Eck 2007; Qualmann and Kessels 2009). (B) Bacterial actin nucleators: VopN and VopF (V. cholerae; Tam et al. 2010); VopL (V. parahaemolyticus, Yu et al. 2011); Tarp (C. trachomatis; Jiwani et al. 2013); BimAma (B. mallei; Benanti et al. 2015), BimAps (B. pseudomallei; Benanti et al. 2015), BimAth (B. thailandensis, Benanti et al. 2015), Sca2 (R. conorii; Madasu et al. 2013); SipC (S. Typhimurium; Hayward and Koronakis 1999); and VipA (L. pneumophila; Franco, Shohdy and Shuman 2012). The number of amino acids of the proteins and the approximate position of regions of interest are indicated (bottom). B, basic region; A, Arp2/3-binding central and acidic motifs; DID/FH3, diaphanous inhibitory domain/Formin homology 3; F, FH1-like domain; D, diaphanous auto-regulatory domain; KIND, kinase noncatalytic C-lobe domain; mFYVE, modified zinc finger motif; NPY, asparagine, proline, tyrosine; VCD, Vop C-terminal domain; NRD, N-terminal repeat domain; CRD, C-terminal repeat domain; AC, auto-chaperone domain; TD, translocator domain. The figure was generated using IBS (Illustrator of Biological Sequences) v1.0 software.