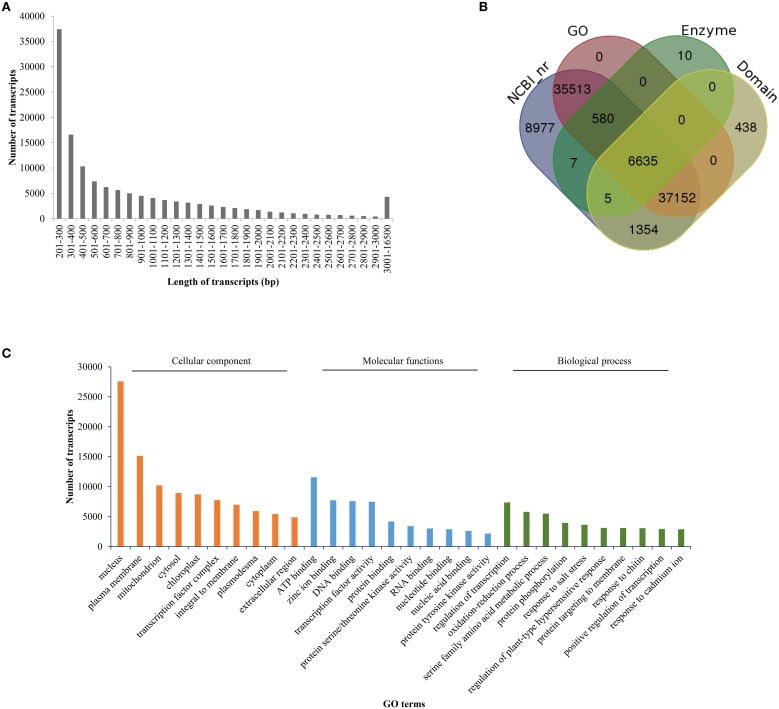
Figure 2.
Size distribution and gene ontology analysis of the assembled transcripts. (A) Assembled transcripts were evaluated for their size distribution and the number of transcripts in each size range is presented. (B) Overlap in the distribution of annotated transcripts based on their databases hits. The LAST search was used to identify transcripts in NCBI nr database whose output was further annotated with Blast2GO for assigning gene ontology terms. PRIAM database and RPS blast were employed for identification of enzymes and conserved domains, respectively. (C) Gene ontology categorization of annotated B. juncea transcripts on the basis of cellular components, molecular functions and biological processes. The numbers of transcripts in the ten most enriched GO terms for each of the categories are shown.