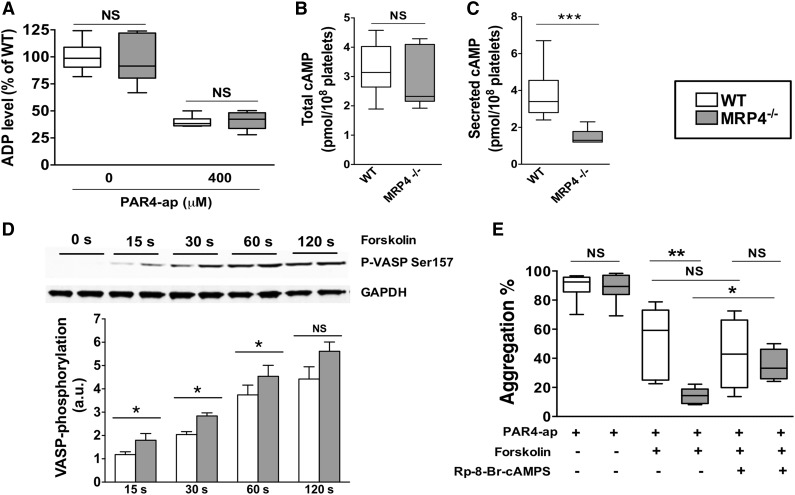
Figure 5.
Role of MRP4 in ADP and cAMP dense-granule storage. (A) ADP was quantified either in lysates of unstimulated or degranulated platelet pellets (obtained by a centrifugation of 2 minutes at 12 000g). Degranulated platelets were obtained after 10 minutes of activation with 400 μM PAR4-ap. Platelets were centrifuged, the supernatant was discarded, and lysis buffer was added to the pellets before quantifying ADP (n = 6). cAMP was quantified in total platelets at rest (n ≥ 6) (B) and in the supernatant of activated platelets (n ≥ 6) (***P < .001) (C). (D) WT and MRP4−/− platelets were incubated with forskolin for the indicated times, and lysates were harvested for western blot analysis and densitometry. Blots were probed with anti P-VASP Ser 157 and loading was controlled with anti-GAPDH. Quantitative analysis is obtained by densitometry using Image J software. Values are mean ± standard error of the mean (n ≥ 4) (*P < .05). Upper panel shows a representative western blot of the phosphorylation kinetics of P-VASP Ser 157 in WT and MRP4−/−. (E) Effect of cAMP-elevating agent and PKA inhibitor on PAR4-ap–induced platelet aggregation. Platelets were preincubated with the PKA inhibitor Rp-8-Br-cAMPS (500 μM) or with vehicle for 10 minutes. Forskolin (5 μM) or vehicle was then added for 15 seconds at 37°C under stirring before activation with PAR4-ap (50 μM) (n ≥ 4) (*P < .05; **P < .001). Boxes represent the interquartile range with median maximal aggregation (horizontal line); whiskers represent the fifth to 95th percentiles. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.