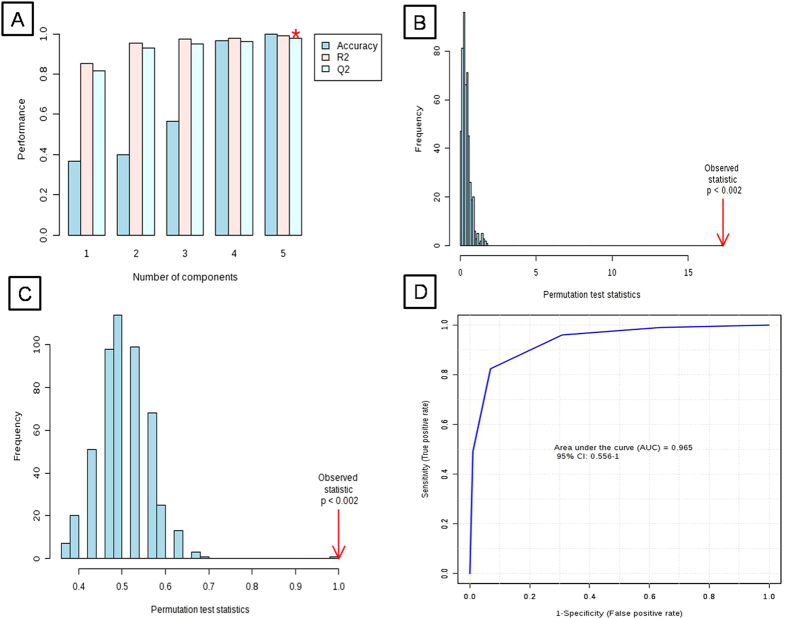
Figure 6.
(A) Cross validation, (B,C) Permutation analysis of PLS-DA models derived from cypermethrin exposed and controls of earthworm Metaphire posthuma tissue extracts. Statistical validation of the PLS-DA by permutation analysis using 500 different model permutations. The goodness of fit and predictive capability of the original class assignments is much higher compared to ratios based on the permutation class assignments. (D) Predictive accuracy of the model discriminating cypermethrin exposed and healthy control earthworms summarised using ROC curve analysis. Area under the curve = 0.965.