Abstract
Background
The temporal relationship between hepatitis B virus (HBV) mutations and hepatocellular carcinoma (HCC) remains unclear.
Methods
We conducted a meta-analysis including cohort and nested case-control studies to prospectively examine the HCC risk associated with common variants of HBV in the PreS, Enhancer II, basal core promoter (BCP) and precore regions. Pertinent studies were identified by searching PubMed, Web of Science and the Chinese Biological Medicine databases through to November 2014. Study-specific risk estimates were combined using fixed or random effects models depending on whether significant heterogeneity was detected.
Results
Twenty prospective studies were identified, which included 8 cohort and 12 nested case-control studies. There was an increased risk of HCC associated with any PreS mutations with a pooled relative risk (RR) of 3.82 [95% confidence interval (CI): 2.59-5.61]. The pooled-RR for PreS deletion was 3.98 (95% CI: 2.28-6.95), which was higher than that of PreS2 start codon mutation (pooled-RR=2.63, 95% CI: 1.30-5.34). C1653T in Enhancer II was significantly associated with HCC risk (pooled-RR=1.83; 95% CI: 1.21-2.76). For mutations in BCP, statistically significant pooled-RRs of HCC were obtained for T1753V (pooled-RR=2.09; 95% CI: 1.49-2.94) and A1762T/G1764A double mutations (pooled-RR=3.11; 95% CI: 2.08-4.64). No statistically significant association with HCC risk was observed for G1896A in the precore region (pooled-RR=0.77; 95% CI: 0.47-1.26).
Conclusions
This study demonstrated that PreS mutations, C1653T, T1753V, and A1762T/G1764A, were associated with an increased risk of HCC. Clinical practices concerning the HCC risk prediction and diagnosis may wish to focus on patients with these mutations.
Keywords: Hepatitis B virus (HBV), mutation, hepatocellular carcinoma (HCC), prospective study, meta-analysis
Introduction
Liver cancer is the fifth most frequently diagnosed cancer and the second leading cause of cancer death worldwide, accounting for almost 6% (782,000) of the total cancer incidence and over 9% (746,000) of the corresponding mortality in 2012. Nearly 80% of the registered cases occur in Eastern Asia and sub-Saharan Africa (1). The prognosis for liver cancer is very poor (overall ratio of mortality to incidence of 0.95) (1), with a 5-year relative survival of less than 20% worldwide (2-4). As such, any promotion of primary prevention, prediction and early diagnosis of liver cancer is a critical public health concern and challenge.
About 80% of hepatocellular carcinoma (HCC) cases are estimated to be associated with hepatitis B virus (HBV) infection, making it one of the most important risk factors in liver carcinogenesis (5). HBV is a partially double-stranded DNA virus that contains four overlapping open reading frames (ORFs): C, the core (nucleocapsid) protein; S, the surface protein; P, viral DNA polymerase (which contains reverse transcriptase activity); and X, the function of which is not totally clear (6). HBV DNA replicates via reverse transcription of an RNA intermediate, but as the reverse transcriptase lacks proofreading function, errors in HBV DNA replication occur at a much higher rate than for other DNA viruses (6). Numerous studies suggest that certain mutations in the HBV genome may be associated with HCC development (7-9). Those mutations, for example, include G1613A (a G-to-A substitution at nucleotide 1613) and C1653T (a C-to-T substitution at nucleotide 1653) in the Enhancer II region, T1753V [a T-to-V (C or A or G) substitution at nucleotide 1753] and the double mutation A1762T/G1764A (an A-to-T substitution at nucleotide 1762 and a G-to-A substitution at nucleotide 1764) at the basal core promoter (BCP) region, G1896A (a G-to-A substitution at nucleotide 1896) and G1899A (a G-to-A substitution at nucleotide 1899) in the precore region (7-29).
However, previous studies vary in the quality of their design and conduct. Most of the designs are hospital-based case-control or cross-sectional studies. They are only able to present a statistical relationship between exposures and the risk of HCC. Longitudinal observations suggest that HBV mutations and HCC may interact or have a reverse pathway, and thus it is possible that certain mutations may occur concurrently or following HCC occurrence (11,22,23). Such a causal bias may produce a spurious association between a specific mutation and HCC risk. As such, prospective studies with a clearly-defined temporal relationship are more convincing than retrospective studies in elucidating potentially causal links between mutation and hepatocarcinogenesis. In addition, considering the prediction value of HBV mutations on HCC occurrence, prospective evaluations could provide more reliable risk estimates, which may guide future clinical practices on HCC screening and early diagnosis. To examine these points further, we therefore conducted a meta-analysis of prospective studies (cohort and nested case-control studies, etc.) to quantitatively evaluate the association between major HBV mutations and HCC risk.
Materials and methods
Study selection
We followed standard criteria for conducting and reporting of meta-analyses of observational studies (30). A comprehensive, computerized literature search was performed using PubMed, Web of Science and the Chinese Biological Medicine through to November, 2014. We searched the databases used the combination of the Medical Subject Heading (MeSH) terms: “hepatitis B virus”, “mutation” and “hepatocellular carcinoma” and the corresponding free terms. The identified publications were reviewed independently for their relevance to the research topic by three authors (YY, JWS and LGZ). We also manually searched the reference lists of relevant publications to identify additional studies. A set of pre-specified inclusion criteria was applied during the review and discrepancies were resolved by consensus. To be included in the meta-analysis, studies had to: (I) use a prospective design; (II) report HBV mutations as the exposure of interest; (III) report HCC or liver cancer as the outcome of interest; and (IV) provide estimates of odds ratios, rate ratios, and hazard ratios with 95% confidence intervals (CIs), or the data necessary to calculate these measures.
Because HCC is a rare outcome in general population, the odds ratio in a nested case-control study is the unbiased estimate of rate ratio or hazard ratio in a cohort study. Thus, we used the relative risk (RR) as a measurement to evaluate association between the HBV mutations and HCC risk in this meta-analysis. If multiple estimates were provided, priority was given to the multivariable adjusted risk estimates which accounted for the major confounding factors in the original studies. Where more than one study was conducted in the same population, we selected either the most recent or most applicable estimates.
Data extraction
We used a standardized protocol and reporting form to abstract the following data from each publication: the first author’s name, the year of publication, the area in which the study was conducted, the duration of follow-up in the original cohort, the age of the study population, the size of the cohort/the number of controls, the number of cases, the detection method, HBV genotype, HBV mutation sites and the corresponding RRs and 95% CIs, and the covariates included for adjustment in multivariable models.
Quality assessment
To assess the study quality, a 9-star system on the basis of the Newcastle-Ottawa Scale was used in which a study was judged on 3 broad perspectives as follows: the selection of study groups, comparability of groups, and ascertainment of either the exposure or outcome of interest for case-control or cohort studies, respectively. The high-quality study was defined as a study with ≥7 awarded stars.
Statistical analysis
To examine the associations between HBV mutations and HCC risk, we used a fixed effect model to pool the study specific estimates unless significant heterogeneity was observed, then the random effect model proposed by DerSimonian and Laird was used, which considered both the within- and between-study variations (31). We conducted subgroup analyses stratified by study design (cohort or nested case-control design), study location (China or other countries), the disease status of controls/cohort [asymptomatic hepatitis B surface antigen carriers (ASC) or chronic hepatitis B (CHB)], and HBV genotypes (C or others).
Sensitivity analyses were further conducted in which one study was removed and the rest were analyzed to evaluate whether the results were affected statistically significantly. We also repeated the analyses in high-quality studies. Heterogeneity among studies was assessed with the Q and the I2 statistic and results were defined as heterogeneous for P<0.10 or I2>50% (32). Small study effects (e.g., publication bias) were evaluated by visual inspection of funnel plots and formal testing by using Egger’s tests (33). When there was an indication of publication bias, we used the trim and fill method to correct for funnel plot asymmetry arising from publication bias (34).
Statistical analyses were conducted using Stata version 13.0 (StataCorp LP, College Station, Texas, USA). Two-sided P<0.05 was considered statistically significant unless otherwise specified.
Results
Literature search and study characteristics
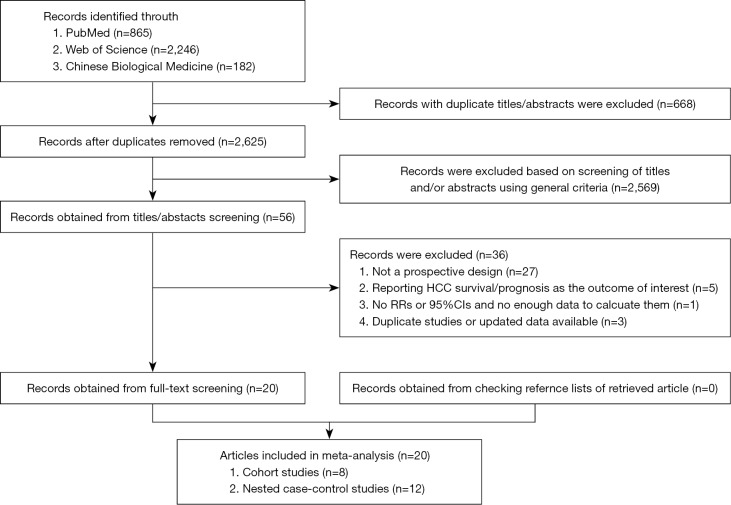
Our systematic literature search yielded a total of 20 articles which prospectively reported the associations between HBV mutations and HCC risk. A flow diagram for the search is presented in Figure 1. Of the 3,293 records identified from the three databases, 668 records were excluded because of duplicated titles and abstracts. After review of the titles and abstracts based on the pre-specified inclusion criteria, 2,569 articles were further excluded. After reviewing full text of the remaining 56 studies, 36 studies were excluded because: (I) studies did not use a prospective design (n=27); (II) studies reported HCC survival or prognosis as the sole outcome of interest (n=5); (III) no RRs or 95% CIs were reported, or there were no enough data to calculate them (n=1); or (IV) newer data were available (n=3). The reference lists of these 36 excluded studies are presented in Supplementary Document S1. Finally, we included 20 prospective studies in the analyses (10-29); and no further studies were identified on checking the reference lists of retrieved articles.
Figure 1.
References searched and selection of studies in the meta-analysis. HCC, hepatocellular carcinoma; RR, relative risk; CI, confidence interval.
Descriptive details of the included studies are summarized in Table 1. There were a total of 1,543 cases in the 20 prospective studies. Eight studies used the entire cohort (12,14,15,19,20,25,26,28) while the remaining 12 used nested case-control designs (10,11,13,16-18,21-24,27,29). Most of the studies were conducted among Chinese populations [10 in Mainland (10,11,13,16,18,19,22-24,27), five in Taiwan area (17,20,21,25,29) and one in Hong Kong (15)], and the others in South Korea (n=2) (12,26), Japan (n=1) (14) and Untied States (n=1) (28). Most of the included studies (n=17) used direct sequencing to detect mutations (10-12,14,15,17-21,23-29), with PreS mutations, C1653T, T1753V, A1762T/G1764A and G1896A, the most frequently reported.
Table 1. Characteristics of studies included in the meta-analysis, 2003-2014.
| References (first author, year) | Study area | Study design | No. of cases | No. of controls/cohorts | Status of controls | Mutation | Detection method | HBV genotype | Adjustment/matching variables |
|---|---|---|---|---|---|---|---|---|---|
| Qu (a), 2014 | China | NCC | 96 | 97 | CHB | Pre-S mutation | Sequence | C | Age, smoking, alcohol drinking |
| Qu (b), 2014 | China | NCC | 152 | 131 | CHB | C1653T, T1753V, A1762T/G1764A, G1896A | Sequence | C | Age, smoking, alcohol drinking |
| Sinn, 2012 | South Korea | Cohort | 24 | 195 | CHB | Pre-S mutation | Sequence | C | Age, sex, HBeAg, cirrhosis, HBV-DNA level |
| Muñoz, 2011 | China | NCC | 345 | 625 | ASC | A1762T/G1764A | Real-time polymerase chain reaction | NA | Age, cohort, screen status and screen test results |
| Kusakabe, 2011 | Japan | Cohort | 13 | 479 | ASC | C1653T, T1753V, A1762T/G1764A, G1896A | Sequence | B, C | Age, sex, body mass index, smoking, alcohol drinking, ALT, c-glutamyl transpeptidase, HBeAg, HBcAg, HBV-DNA, Genotype |
| Yuen, 2009 | Hong Kong, China | Cohort | 40 | 820 | CHB | A1762T/G1764A | Sequence | B, C | Age, gender, HBV genotype, core promoter and precore mutations, HBeAg/anti-HBe status, HBV DNA, ALT levels and LC |
| Yuan, 2009 | China | NCC | 50 | 106 | ASC | A1762T/G1764A | Real-time polymerase chain reaction | NA | Age, years between blood draw and measurement of HBV DNA double mutation, neighborhood of residence at recruitment, cigarette smoking, heavy alcohol consumption, and serum concentration of retinol |
| Sung, 2009 | Taiwan, China | NCC | 116 | 154 | ASC | A1762T/G1764A, G1896A | Sequence | A, B, C | OR and 95% CI calculated on the distribution of cases and controls according to the mutations. |
| Fang (a), 2008 | China | NCC | 33 | 33 | ASC | Pre-S mutations | Sequence | B, C | Age, sex and status of core promoter sequence, HBeAg, Anti-HBe, genotype, ALT |
| Fang (b), 2008 | China | Cohort | 61 | 2258 | ASC | C1653T, T1753V, A1762T/G1764A | Sequence | NA | Age, sex. HBeAg and abnormal ALT were not confounders in this study. |
| Yang, 2008 | Taiwan, China | Cohort | 153 | 2762 | ASC | A1762T/G1764A, G1896A, Pre-S mutation (Unpublished data) | Sequence | B, C | Age, sex, cigarette smoking, alcohol drinking, ALT level, LC, HBV DNA level, HBV genotype |
| Chou, 2008 | Taiwan, China | NCC | 132 | 204 | ASC | T1753V, A1762T/G1764A, G1896A | Sequence | A, B, C | OR and 95% CI calculated on the distribution of cases and controls according to the mutations. |
| Cao, 2008 | China | NCC | 47 | 50 | CHB | Pre S mutations | Other | NA | OR and 95% CI calculated on the distribution of cases and controls according to the mutations. |
| Guo, 2008 | China | NCC | 58 | 71 | CHB | C1653T, T1753V, A1762T/G1764A | Sequence | B, C | OR and 95% CI calculated on the distribution of cases and controls according to the mutations. |
| Zhu, 2008 | China | NCC | 20 | 83 | CHB | A1762T/G1764A | Sequence | C | OR and 95% CI calculated on the distribution of cases and controls according to the mutations. |
| Chen, 2007 | Taiwan, China | Cohort | 7 | 141 | ASC, CHB | Pre-S deletions, G1896A | Sequence | B, C | Age, sex, ALT, total bilirubin, HBV-DNA, HBV genotypes |
| Jang, 2007 | South Korea | Cohort | 6 | 29 | ASC, CHB, LC | A1762T/G1764A | Sequence | C | OR and 95% CI calculated on the distribution of cases and controls according to the mutations. |
| Zhang, 2007 | China | NCC | 32 | 32 | CHB | A1762T/G1764A | Sequence | NA | Age sex, occupation, living environment |
| Tong, 2006 | United States | Cohort | 31 | 400 | CHB | A1762T/G1764A, G1896A | Sequence | A, B, C | OR and 95% CI calculated on the distribution of cases and controls according to the mutations. |
| Kao, 2003 | Taiwan, China | NCC | 127 | 123 | CHB | A1762T/G1764A | Sequence | B, C | Age, sex, LC, genotype |
NCC, nested case-control study; ASC, asymptomatic hepatitis B surface antigen carriers; CHB, chronic hepatitis B; LC, liver cirrhosis; HBeAg, hepatitis B e antigen; ALT, alanine aminotransferase; NA, not available; other, restriction fragment length polymorphism and polyacrylamide gel electrophoresis.
Study-specific quality scores are summarized in Tables S1 and S2. The ranges of quality scores were from 4 to 9 for cohort studies and 5 to 9 for nested case-control studies; and the median score was 7 for both study designs. Six cohort and nine nested case-control studies were awarded ≥7 stars and thus defined as high-quality studies.
PreS mutations and HCC risk
The pooled-RRs of HCC for any PreS mutations are shown in Figure 2 and Table 2. All of these included studies are defined as high-quality studies. Compared with individuals without any PreS mutations, the pooled-RR was 3.82 (95% CI: 2.59-5.61) for those with the mutations. No statistical heterogeneity was detected (Pheterogeneity=0.340, I2=11.7%). The risk was higher in Chinese population (pooled-RR=4.02; 95% CI: 2.62-6.17). Concerning the two main mutation types in PreS region, the pooled RR for PreS deletions was 3.98 (95% CI: 2.28-6.95; I2=9.2%, Pheterogeneity=0.347), which was higher than the PreS2 start codon mutation (pooled-RR=2.63, 95% CI: 1.30-5.34; I2=0.0%, Pheterogeneity=0.673). There was no evidence of publication bias for PreS mutations as tested using Egger’s test (P=0.139) and by visual inspection of funnel plot (Figure S1).
Figure 2.
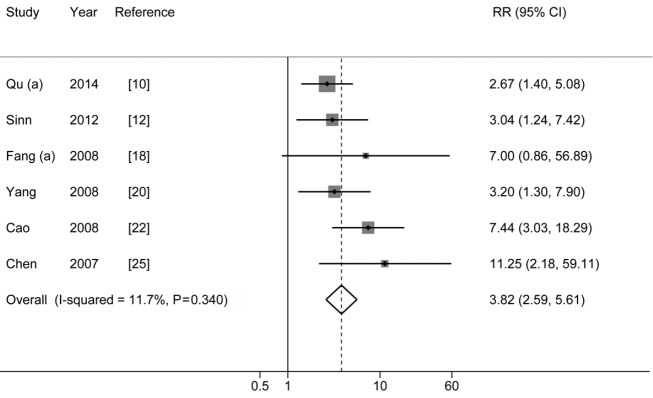
Results from meta-analysis of association between PreS mutations and hepatocellular carcinoma risk.
Table 2. Summary estimates and corresponding 95% CI for hepatitis B virus mutations and hepatocellular carcinoma.
| Mutations | No. of studies | Pooled RR | 95% CI | I2 (%) | P for heterogeneity |
|---|---|---|---|---|---|
| Any PreS mutations† | 6 | 3.82 | 2.59-5.61 | 11.7 | 0.340 |
| PreS deletions | 4 | 3.98 | 2.28-6.95 | 9.2 | 0.347 |
| PreS2 start codon mutation | 3 | 2.63 | 1.30-5.34 | 0 | 0.673 |
| C1653T | 4 | 1.83 | 1.21-2.76 | 0 | 0.950 |
| T1753V | 5 | 2.09 | 1.49-2.94 | 0 | 0.933 |
| A1762T/G1764A | 15 | 3.11 | 2.08-4.64 | 78.1 | <0.001 |
| G1896A | 7 | 0.77 | 0.47-1.26 | 75.3 | <0.001 |
CI, confidence interval; RR, relative risk; †, any PreS mutations include any point mutations or deletions occurred in PreS1 or PreS2 regions.
C1653T and HCC risk
As shown in Figure 3 and Table 2, we observed a statistically significant association between C1653T and HCC risk (pooled-RR=1.83; 95% CI: 1.21-2.76). No statistical heterogeneity was detected (Pheterogeneity=0.950, I2=0.0%). Compared with studies with CHB patients as control group (pooled-RRCHB=1.75, 95% CI: 1.11-2.76), the strength of association was stronger when we used ASC as control subjects (pooled-RRASC=2.20, 95% CI: 0.85-5.72). When we excluded one study conducted in Japan, the risk estimate for the Chinese population was similar to the overall estimate. When we excluded one low-quality study, the results did not alter materially. There was no evidence of publication bias as tested using Egger’s test (P=0.487) and by visual inspection of funnel plot (Figure S2).
Figure 3.
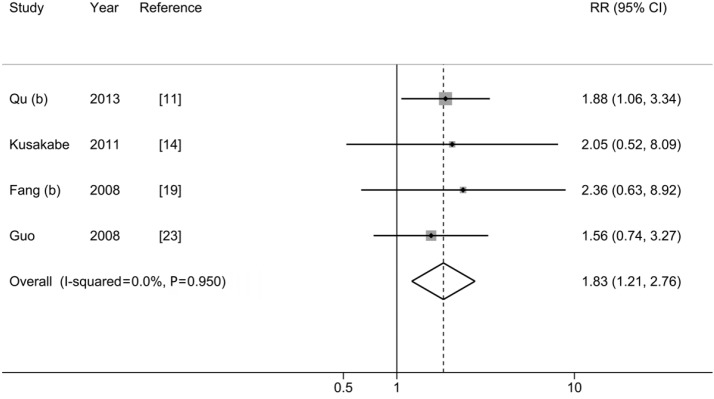
Results from meta-analysis of association between C1653T and hepatocellular carcinoma risk.
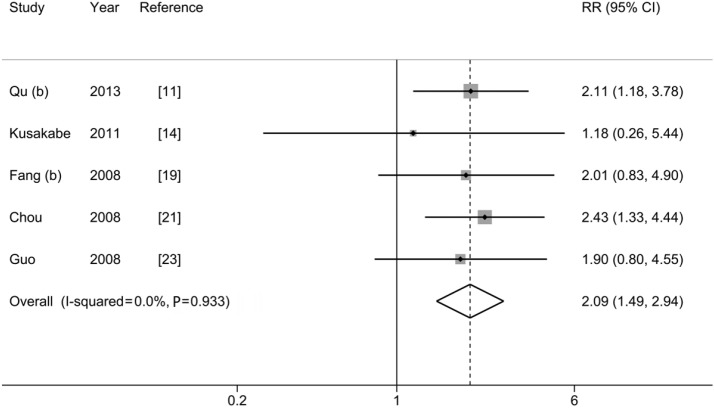
T1753V and HCC risk
Figure 4 and Table 2 show the pooled-RR of HCC for T1753V. There was a significant increase in HCC risk for individuals with T1753V compared with those without the mutation (RR=2.09; 95% CI: 1.49-2.94; Pheterogeneity=0.933, I2=0%). The result obtained from cohort studies (pooled-RR=1.76; 95% CI: 0.82-3.78; Pheterogeneity=0.533, I2=0%) was lower than that from the nested case-control studies (pooled-RR=2.19; 95% CI: 1.50-3.19; Pheterogeneity=0.890, I2=0%). The risk for studies with ASC as control subjects (pooled-RRASC=2.15; 95% CI: 1.34-3.45; Pheterogeneity=0.678, I2=0%) was slightly higher than that of CHB patients as controls (pooled-RRCHB=2.04; 95% CI: 1.26-3.31; Pheterogeneity=0.844, I2=0%). When we excluded the Japanese study, the strength of the association among the Chinese population slightly increased (pooled-RR=2.16, 95% CI: 1.53-3.05; Pheterogeneity=0.967, I2=0%). When we repeated the analysis in three high-quality studies, the pooled-RR did not change materially. Evidence of publication bias was detected using Egger’s test (P=0.037) but not Begg’s test (P =0.221) (Figure S3), and the overall estimate did not change when we used the trim and fill method to correct for funnel plot asymmetry arising from publication bias.
Figure 4.
Results from meta-analysis of association between T1753V and hepatocellular carcinoma risk.
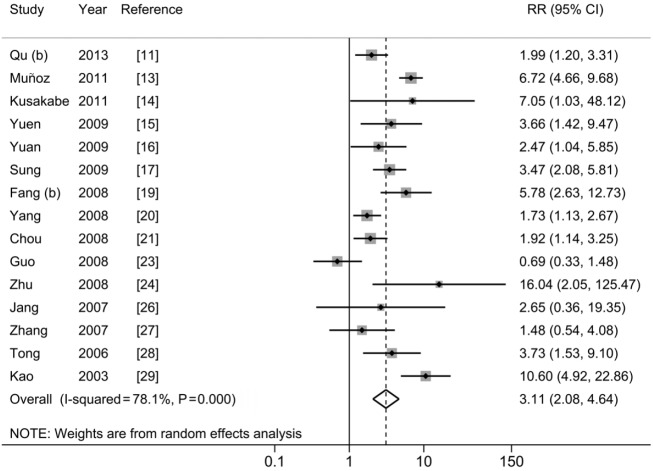
A1762T/G1764A and HCC risk
The pooled-RR of HCC for A1762T/G1764A is shown in Figure 5 and Table 2. A total of 15 studies reported the association between A1762T/G1764A double mutations and HCC risk. Individuals with these double mutations were at higher risk of developing HCC (pooled-RR=3.11; 95% CI: 2.08-4.64; Pheterogeneity<0.001, I2=78.1%) than those without these mutations. The pooled-RR derived from cohort studies (pooled-RR=3.27; 95% CI: 1.93-5.55; Pheterogeneity=0.089, I2=47.6%) was slightly higher than that from the nested case-control studies (pooled-RR=2.96; 95% CI: 1.69-5.19; Pheterogeneity<0.001, I2=85.0%). When we stratified by the disease status among controls, the pooled-RR with ASC as control subjects was 3.34 (95% CI: 2.01-5.57; Pheterogeneity<0.001, I2=80.2%), higher than that of CHB patients as controls (pooled-RR=2.96; 95% CI: 1.41-6.21; Pheterogeneity<0.001, I2=80.8%). Moreover, compared with other study areas (Japan, South Korea and US), the pooled-RR was higher for studies conducted in China (RR=3.00; 95% CI: 1.92-4.70; Pheterogeneity<0.001, I2=82.6%). When we stratified by study quality, the pooled-RR of 10 high-quality studies was 3.48 (95% CI: 1.90-6.37; Pheterogeneity<0.001, I2=84.0%), which was higher than that of 5 low-quality studies (pooled-RR=2.48; 95% CI: 1.88-3.28; Pheterogeneity=0.394, I2=2.3%). There was no evidence of publication bias based on Egger’s test (P=0.945) and by visual inspection of funnel plot (Figure S4).
Figure 5.
Results from meta-analysis of association between A1762T/G1764A and hepatocellular carcinoma risk.
G1896A and HCC risk
As shown in Figure 6 and Table 2, no statistically significant association was observed between G1896A and HCC risk (pooled-RR=0.77; 95% CI: 0.47-1.26; Pheterogeneity<0.001, I2=75.3%). Of note, there was a statistically significant inverse association among Chinese population (pooled-RR=0.60, 95% CI: 0.38-0.95; Pheterogeneity=0.012, I2=68.9%). In contrast, an increased risk (pooled-RR=2.06, 95% CI: 1.09-3.89; Pheterogeneity<0.001, I2=77.1%) was observed for studies conducted in other countries (Japan and United States). The pooled-RR derived from cohort studies (pooled-RR=0.74; 95% CI: 0.21-2.62; Pheterogeneity<0.001, I2=86.1%) was slightly lower than that from the nested case-control studies (pooled-RR=0.80; 95% CI: 0.61-1.05; Pheterogeneity=0.411, I2=0%). When we stratified by study quality, the pooled-RR of 3 high-quality studies was 0.46 (95% CI: 0.14-1.56; Pheterogeneity=0.08, I2=60.4%), which was lower than that of 4 low-quality studies (pooled-RR=0.96; 95% CI: 0.63-1.45; Pheterogeneity=0.05, I2=61.5%). No indication of publication bias was found via Egger’s test (P=0.840) and by visual inspection of funnel plot (Figure S5).
Figure 6.
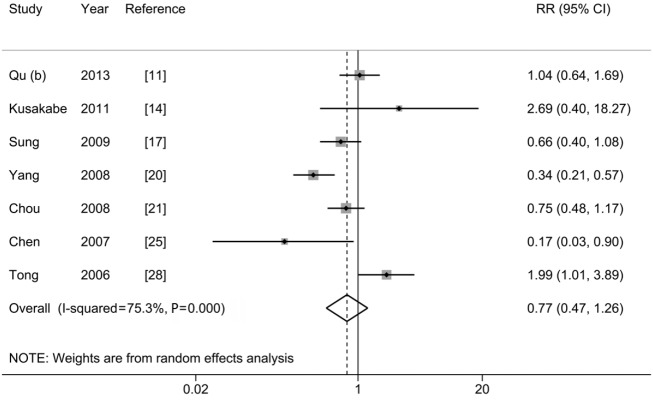
Results from meta-analysis of association between G1896A and hepatocellular carcinoma risk.
In sensitivity analyses, we sequentially excluded one study to recalculate the pooled-RRs of HCC for these HBV-specific mutations. The pooled-RRs were not altered materially in comparison with their corresponding overall estimates (data not shown).
Discussion
This meta-analysis systematically evaluated major HBV mutations in association with the HCC risk using prospective epidemiological studies. We found that PreS mutations, C1653T, T1753V, and A1762T/G1764A, were statistically significantly associated with an increased risk of HCC. The strength of association ranked the highest for PreS mutations, followed by A1762T/G1764A double mutations. The summarized risk estimates were robust across subgroup and sensitivity analyses.
It is biologically plausible that certain HBV mutants in essential genes such as PreS, Enhancer II and BCP may contribute to hepatocarcinogenesis. Several specific mutations in the PreS gene may induce an unbalanced production of envelope proteins that accumulate in the endoplasmic reticulum of the hepatocytes, and may activate the endoplasmic reticulum stress signaling pathways with consequent induction of oxidative DNA damage and genomic instability (35). The cytotoxic effects exerted by the intracellular accumulation of surface proteins can contribute to liver damage, favoring the progression of the disease toward cirrhosis and the development of HCC (35). It has been proposed that several ubiquitous and liver-specific trans-regulating nuclear factors such as transcription factor Sp1, CCAAT/enhancer-binding protein and HNF4 could bind HBV at nucleotides 1653, 1753, and 1762/1764 (36-38). These HBV sites overlap with the X gene—a pivotal ORF encodes the X protein. Mutations at these sites such as C1653T, T1753V, and A1762T/G1764A may alter the binding ability of the transregulating nuclear factors which is responsible for liver carcinogenesis (38). Those mutations may also lead to amino acid alterations of X protein, and such alterations in the X protein have been suggested to transactivate oncogenes which could contribute to the hepatocarcinogenesis (39).
Three previous meta-analyses have summarized the published studies and provided pooled risk estimates for different HBV mutations sites in association with HCC risk (7-9). Compared to the previous meta-analysis which reported a 46% increased risk of HCC in HBV-infected patients with precore mutation G1896A (8), we did not observe such significant association in present meta-analysis. Of note, for studies conducted in China, G1896A was inversely associated with HCC in our study (pooled-RR=0.60, 95% CI: 0.38-0.95). Furthermore, the strength of the association for C1653T, T1753V, and A1762T/G1764A in core promoter regions all attenuated in our study in comparison with previous meta-analyses. These differences in strength of association may be due to the inclusion of large numbers of retrospective or cross-sectional studies in previous meta-analyses, potentially introducing various biases, such as inverse casual and selection biases. For example, since some mutations may occur concurrently or after HCC diagnosis, a spurious causal relationship may be observed using cross-sectional or retrospective case-control designs. Over four-fifths of all previous studies assessing the association between HBV mutations and HCC risk used a hospital-based case-control design, exposing the pooled estimates to these biases. Moreover, the heterogeneity of the risk estimates between our study and the previous meta-analyses may be partly due to differences in the study location, study population, HBV genotypes etc. For example, most of studies in our meta-analysis are from China with an epidemic of the genotypes B and C, whereas previous meta-analysis included more studies conducted in other population or countries with other HBV genotypes. In addition, differences in adjustment for potential confounders may also account for the heterogeneity of the results between our study and previous meta-analyses.
This study has several limitations. Firstly, most studies included in this meta-analysis were conducted in Eastern Asia, where HBV genotypes B and C are dominant (40). Thus, generalization of these results to other populations with a different distribution of HBV genotypes should be interpreted with caution. Secondly, because of the nature of the observational design and the limitations of the meta-analysis approach to eliminate potential biases inherent in the original studies, selection bias and residual confounding may distort the association between HBV mutations and HCC. For example, HBV mutation data were not available for all participants because HBV DNA could not be efficiently amplified by polymerase chain reaction if the viral load was below the detection limit. Thus, a selection bias may have been introduced given HBV mutation data were more likely to be obtained from the participants with a high viral load than those with lower viral loads. In addition, some studies eligible for inclusion did not adjust or consider viral load or acceptance of the antiviral treatments in their analyses; thus the results may be biased by the residual confounding effects of these factors. Thirdly, there was significant heterogeneity for the results for some mutation sites. The observed between-study heterogeneity may result from various sources, for example, across studies, experimental conditions or detection methods may be different, the size of cohort and the length of follow-up may vary, or other modifiers or confounders such as HBV genotype and HBV DNA load may be present. Given the small number of included studies, we failed to carry out meta-regression analyses to further explore the sources of heterogeneity. Finally, publication bias may influence the results. Although there is little evidence of publication bias across mutation sites in this meta-analysis, tests for publication bias have low statistical power, especially when the number of studies is limited.
Conclusions
This study provides evidence that PreS mutations, C1653T, T1753V, and A1762T/G1764A, are associated with an increased risk of HCC. Future studies should place emphasis on the potential mechanisms to further elucidate the possibly causal links underlying these observed associations. Clinical practice concerning HCC risk prediction and diagnosis may wish to focus on patients with these mutations.
Acknowledgements
Funding: This work was supported by the fund of the National Key Basic Research Program “973 project” (2015CB554000), and grants from the State Key Project Specialized for Infectious Diseases of China (2008ZX10002-015 and 2012ZX10002008-002).
Document S1
The reference lists of the 36 excluded studies. Studies were excluded because: (I) studies (41-50) did not use a prospective design (n=27) (51-67); (II) studies reported HCC survival or prognosis as the sole outcome of interest (n=5) (68-72); (III) no relative risks (RRs) or 95% confidence intervals (CIs) were reported, or there were not enough data to calculate them (n=1) (73); or (IV) newer data were available (n=3) (74-76).
Table S1. Methodological quality of cohort studies included in the meta-analysis*.
| Reference (first author, year) | Representativeness of exposed cohort | Selection of unexposed cohort | Ascertainment of exposure | Outcome of interest not present at start of study | Control for important factor or additional factor† | Outcome assessment | Follow-up long enough for outcomes to occur‡ | Adequacy of follow-up of cohorts | Total quality scores |
|---|---|---|---|---|---|---|---|---|---|
| Sinn, 2012 | – | ☆ | ☆ | ☆ | ☆☆ | ☆ | – | ☆ | 7 |
| Kusakabe, 2011 | ☆ | ☆ | ☆ | ☆ | ☆☆ | ☆ | ☆ | ☆ | 9 |
| Yuen, 2009 | – | ☆ | ☆ | ☆ | ☆☆ | ☆ | – | ☆ | 7 |
| Fang (b), 2008 | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | – | ☆ | 7 |
| Yang, 2008 | ☆ | ☆ | ☆ | ☆ | ☆☆ | ☆ | ☆ | ☆ | 9 |
| Chen, 2007 | – | ☆ | ☆ | ☆ | ☆☆ | ☆ | ☆ | ☆ | 8 |
| Jang, 2007 | – | ☆ | ☆ | – | – | ☆ | ☆ | ☆ | 5 |
| Tong, 2006 | – | ☆ | ☆ | ☆ | – | ☆ | – | – | 4 |
*, a study could be awarded a maximum of one star for each item except for the item Control for important factor or additional factor; †, a maximum of 2 stars could be awarded for this item. Studies that matched or controlled for age and sex received one star, whereas studies that controlled for more than 3 of other important confounders such as alcohol drinking, smoking, HBV DNA, Genotype, HBeAg, ALT, liver cirrhosis received an additional star; ‡, a cohort study with a follow-up time >10 year was assigned one star.
Table S2. Methodological quality of nested case-control studies included in the meta-analysis*.
| Reference (first author, year) | Adequate definition of cases | Representativeness of cases | Selection of control subjects | Definition of control subjects | Control for important factor or additional factor† | Exposure assessment | Same method of ascertainment for all subjects | Nonresponse rate‡ | Total quality scores |
|---|---|---|---|---|---|---|---|---|---|
| Qu (a), 2014 | ☆ | – | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | 7 |
| Qu (b), 2014 | ☆ | – | ☆ | ☆ | ☆ | ☆ | ☆ | – | 6 |
| Munoz, 2011 | ☆ | – | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | 7 |
| Yuan, 2009 | ☆ | ☆ | ☆ | ☆ | ☆☆ | ☆ | ☆ | ☆ | 9 |
| Sung, 2009 | ☆ | – | ☆ | ☆ | – | ☆ | ☆ | – | 5 |
| Fang (a), 2008 | ☆ | – | ☆ | ☆ | ☆☆ | ☆ | ☆ | ☆ | 8 |
| Chou, 2008 | ☆ | – | ☆ | ☆ | – | ☆ | ☆ | – | 5 |
| Cao, 2008 | ☆ | ☆ | ☆ | ☆ | – | ☆ | ☆ | ☆ | 7 |
| Guo, 2008 | ☆ | ☆ | ☆ | ☆ | – | ☆ | ☆ | ☆ | 7 |
| Zhu, 2008 | ☆ | ☆ | ☆ | ☆ | – | ☆ | ☆ | ☆ | 7 |
| Zhang, 2007 | – | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | 7 |
| Kao, 2003 | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | 8 |
*, a study could be awarded a maximum of one star for each item except for the item Control for important factor or additional factor; †, a maximum of 2 stars could be awarded for this item. Studies that matched or controlled for age and sex received one star, whereas studies that controlled for more than 3 of other important confounders such as alcohol drinking, smoking, HBV DNA, Genotype, HBeAg, ALT, liver cirrhosis received an additional star; ‡, a One star was assigned if there was no significant difference in the response rate between control subjects and cases by using the chi-square test (P<0.05).
Figure S1.
Funnel plot for the association between PreS mutations and hepatocellular carcinoma risk. SE, standard error; RR, relative risk.
Figure S2.
Funnel plot for the association between C1653T and hepatocellular carcinoma risk. SE, standard error; RR, relative risk.
Figure S3.
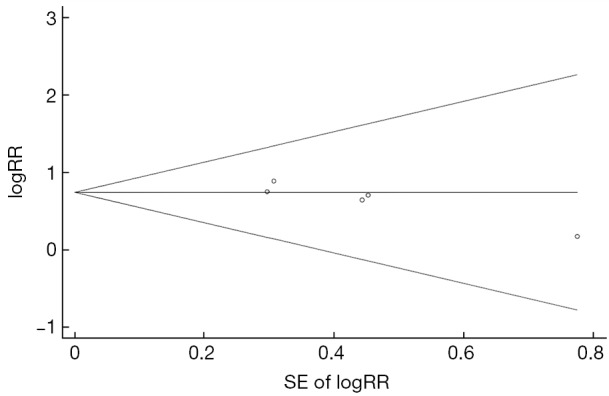
Funnel plot for the association between T1753V and hepatocellular carcinoma risk. SE, standard error; RR, relative risk.
Figure S4.
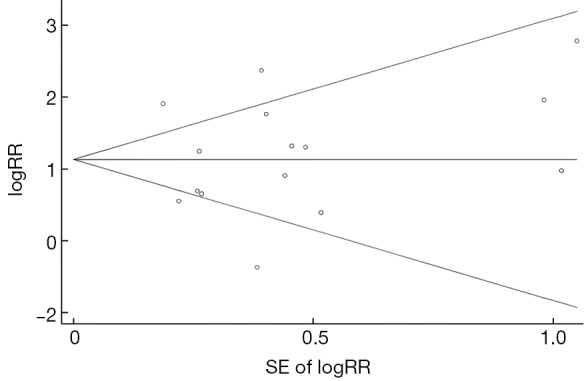
Funnel plot for the association between A1762T/G1764A and hepatocellular carcinoma risk. SE, standard error; RR, relative risk.
Figure S5.
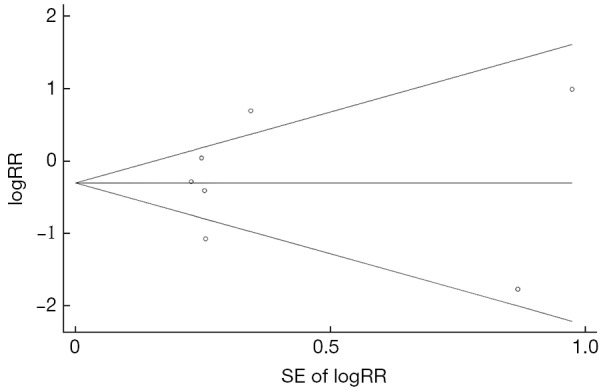
Funnel plot for the association between G1896A and hepatocellular carcinoma risk. SE, standard error; RR, relative risk.
Footnotes
Conflicts of Interest: The authors have no conflict of interest to declare.
References
- 1.Ferlay J, Soerjomataram I, Ervik M, et al. GLOBOCAN 2012 v1.0, cancer incidence and mortality worldwide: IARC CancerBase No. 11. Lyon: IARC, 2013. Available online: http://globocan.iarc.fr. Accessed on 1/11/2014.
- 2.Zeng H, Zheng R, Guo Y, et al. Cancer survival in China, 2003-2005: a population-based study. Int J Cancer 2015;136:1921-30. [DOI] [PubMed] [Google Scholar]
- 3.De Angelis R, Sant M, Coleman MP, et al. Cancer survival in Europe 1999-2007 by country and age: results of EUROCARE--5-a population-based study. Lancet Oncol 2014;15:23-34. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5-29. [DOI] [PubMed] [Google Scholar]
- 5.El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 2012;142:1264-1273.e1. [DOI] [PMC free article] [PubMed]
- 6.Chotiyaputta W, Lok AS. Hepatitis B virus variants. Nat Rev Gastroenterol Hepatol 2009;6:453-62. [DOI] [PubMed] [Google Scholar]
- 7.Liu S, Zhang H, Gu C, et al. Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: a meta-analysis. J Natl Cancer Inst 2009;101:1066-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liao Y, Hu X, Chen J, et al. Precore mutation of hepatitis B virus may contribute to hepatocellular carcinoma risk: evidence from an updated meta-analysis. PLoS One 2012;7:e38394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi HP, Zhang J, Shang XC, et al. Hepatitis B virus gene C1653T polymorphism mutation and hepatocellular carcinoma risk: an updated meta-analysis. Asian Pac J Cancer Prev 2013;14:1043-7. [DOI] [PubMed] [Google Scholar]
- 10.Qu LS, Liu JX, Liu TT, et al. Association of hepatitis B virus pre-S deletions with the development of hepatocellular carcinoma in Qidong, China. PLoS One 2014;9:e98257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu LS, Zhu J, Liu TT, et al. Effect of combined mutations in the enhancer II and basal core promoter of hepatitis B virus on development of hepatocellular carcinoma in Qidong, China. Hepatol Res 2014;44:1186-95. [DOI] [PubMed] [Google Scholar]
- 12.Sinn DH, Choi MS, Gwak GY, et al. Pre-s mutation is a significant risk factor for hepatocellular carcinoma development: a long-term retrospective cohort study. Dig Dis Sci 2013;58:751-8. [DOI] [PubMed] [Google Scholar]
- 13.Muñoz A, Chen JG, Egner PA, et al. Predictive power of hepatitis B 1762T/1764A mutations in plasma for hepatocellular carcinoma risk in Qidong, China. Carcinogenesis 2011;32:860-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kusakabe A, Tanaka Y, Inoue M, et al. A population-based cohort study for the risk factors of HCC among hepatitis B virus mono-infected subjects in Japan. J Gastroenterol 2011;46:117-24. [DOI] [PubMed] [Google Scholar]
- 15.Yuen MF, Tanaka Y, Fong DY, et al. Independent risk factors and predictive score for the development of hepatocellular carcinoma in chronic hepatitis B. J Hepatol 2009;50:80-8. [DOI] [PubMed] [Google Scholar]
- 16.Yuan JM, Ambinder A, Fan Y, et al. Prospective evaluation of hepatitis B 1762(T)/1764(A) mutations on hepatocellular carcinoma development in Shanghai, China. Cancer Epidemiol Biomarkers Prev 2009;18:590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sung FY, Jung CM, Wu CF, et al. Hepatitis B virus core variants modify natural course of viral infection and hepatocellular carcinoma progression. Gastroenterology 2009;137:1687-97. [DOI] [PubMed] [Google Scholar]
- 18.Fang ZL, Sabin CA, Dong BQ, et al. Hepatitis B virus pre-S deletion mutations are a risk factor for hepatocellular carcinoma: a matched nested case-control study. J Gen Virol 2008;89:2882-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang ZL, Sabin CA, Dong BQ, et al. HBV A1762T, G1764A mutations are a valuable biomarker for identifying a subset of male HBsAg carriers at extremely high risk of hepatocellular carcinoma: a prospective study. Am J Gastroenterol 2008;103:2254-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang HI, Yeh SH, Chen PJ, et al. Associations between hepatitis B virus genotype and mutants and the risk of hepatocellular carcinoma. J Natl Cancer Inst 2008;100:1134-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chou YC, Yu MW, Wu CF, et al. Temporal relationship between hepatitis B virus enhancer II/basal core promoter sequence variation and risk of hepatocellular carcinoma. Gut 2008;57:91-7. [DOI] [PubMed] [Google Scholar]
- 22.Cao Z, Bai X, Guo X, et al. High prevalence of hepatitis B virus pre-S mutation and its association with hepatocellular carcinoma in Qidong, China. Arch Virol 2008;153:1807-12. [DOI] [PubMed] [Google Scholar]
- 23.Guo X, Jin Y, Qian G, et al. Sequential accumulation of the mutations in core promoter of hepatitis B virus is associated with the development of hepatocellular carcinoma in Qidong, China. J Hepatol 2008;49:718-25. [DOI] [PubMed] [Google Scholar]
- 24.Zhu R, Zhang HP, Yu H, et al. Hepatitis B virus mutations associated with in situ expression of hepatitis B core antigen, viral load and prognosis in chronic hepatitis B patients. Pathol Res Pract 2008;204:731-42. [DOI] [PubMed] [Google Scholar]
- 25.Chen CH, Hung CH, Lee CM, et al. Pre-S deletion and complex mutations of hepatitis B virus related to advanced liver disease in HBeAg-negative patients. Gastroenterology 2007;133:1466-74. [DOI] [PubMed] [Google Scholar]
- 26.Jang JW, Lee YC, Kim MS, et al. A 13-year longitudinal study of the impact of double mutations in the core promoter region of hepatitis B virus on HBeAg seroconversion and disease progression in patients with genotype C chronic active hepatitis. J Viral Hepat 2007;14:169-75. [DOI] [PubMed] [Google Scholar]
- 27.Zhang F, Shao YF, Gao JD, et al. Risk features of HBV in human hepatocarcinogenesis: a nested case-controlled study. Zhonghua Wai Ke Za Zhi 2007;45:1482-4. [PubMed] [Google Scholar]
- 28.Tong MJ, Blatt LM, Kao JH, et al. Precore/basal core promoter mutants and hepatitis B viral DNA levels as predictors for liver deaths and hepatocellular carcinoma. World J Gastroenterol 2006;12:6620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kao JH, Chen PJ, Lai MY, et al. Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology 2003;124:327-34. [DOI] [PubMed] [Google Scholar]
- 30.Stroup DF, Berlin JA, Morton SC, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA 2000;283:2008-12. [DOI] [PubMed] [Google Scholar]
- 31.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986;7:177-88. [DOI] [PubMed] [Google Scholar]
- 32.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539-58. [DOI] [PubMed] [Google Scholar]
- 33.Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duval S, Tweedie R. A nonparametric “trim and fill” method of accounting for publication bias in meta-analysis. JASA 2000;95:89-98. [DOI] [PubMed] [Google Scholar]
- 35.Pollicino T, Cacciola I, Saffioti F, et al. Hepatitis B virus PreS/S gene variants: pathobiology and clinical implications. J Hepatol 2014;61:408-17. [DOI] [PubMed] [Google Scholar]
- 36.López-Cabrera M, Letovsky J, Hu KQ, et al. Multiple liver-specific factors bind to the hepatitis B virus core/pregenomic promoter: trans-activation and repression by CCAAT/enhancer binding protein. Proc Natl Acad Sci U S A 1990;87:5069-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Ou JH. Differential regulation of hepatitis B virus gene expression by the Sp1 transcription factor. J Virol 2001;75:8400-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quarleri J. Core promoter: a critical region where the hepatitis B virus makes decisions. World J Gastroenterol 2014;20:425-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng SA, Lee C. Hepatitis B virus X gene and hepatocarcinogenesis. J Gastroenterol 2011;46:974-90. [DOI] [PubMed] [Google Scholar]
- 40.Sunbul M. Hepatitis B virus genotypes: global distribution and clinical importance. World J Gastroenterol 2014;20:5427-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 41.Park YM, Jang JW, Yoo SH, et al. Combinations of eight key mutations in the X/preC region and genomic activity of hepatitis B virus are associated with hepatocellular carcinoma. J Viral Hepat 2014;21:171-7. [DOI] [PubMed] [Google Scholar]
- 42.Araujo OC, Barros JJ, do Ó KM, et al. Genetic variability of hepatitis B and C viruses in Brazilian patients with and without hepatocellular carcinoma. J Med Virol 2014;86:217-23. [DOI] [PubMed] [Google Scholar]
- 43.Xie J, Zhang Y, Zhang Q, et al. Interaction of signal transducer and activator of transcription 3 polymorphisms with hepatitis B virus mutations in hepatocellular carcinoma. Hepatology 2013;57:2369-77. [DOI] [PubMed] [Google Scholar]
- 44.Wang H, Gao CF. Hepatocellular carcinoma risk and hepatitis B virus mutations. Zhonghua Gan Zang Bing Za Zhi (in Chinese) 2013;21:788-92. [PubMed] [Google Scholar]
- 45.Thongbai C, Sa-nguanmoo P, Kranokpiruk P, et al. Hepatitis B virus genetic variation and TP53 R249S mutation in patients with hepatocellular carcinoma in Thailand. Asian Pac J Cancer Prev 2013;14:3555-9. [DOI] [PubMed] [Google Scholar]
- 46.Khan A, Al Balwi MA, Tanaka Y, et al. Novel point mutations and mutational complexes in the enhancer II, core promoter and precore regions of hepatitis B virus genotype D1 associated with hepatocellular carcinoma in Saudi Arabia. Int J Cancer 2013;133:2864-71. [DOI] [PubMed] [Google Scholar]
- 47.Gopalakrishnan D, Keyter M, Shenoy KT, et al. Hepatitis B virus subgenotype A1 predominates in liver disease patients from Kerala, India. World J Gastroenterol 2013;19:9294-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee MH, Kim do Y, Kim JK, et al. Combination of preS deletions and A1762T/G1764A mutations in HBV subgenotype C2 increases the risk of developing HCC. Intervirology 2012;55:296-302. [DOI] [PubMed] [Google Scholar]
- 49.Kao JH, Liu CJ, Jow GM, et al. Fine mapping of hepatitis B virus pre-S deletion and its association with hepatocellular carcinoma. Liver Int 2012;32:1373-81. [DOI] [PubMed] [Google Scholar]
- 50.Jang JW, Chun JY, Park YM, et al. Mutational complex genotype of the hepatitis B virus X/precore regions as a novel predictive marker for hepatocellular carcinoma. Cancer Sci 2012;103:296-304. [DOI] [PubMed] [Google Scholar]
- 51.Chu CM, Lin CC, Lin SM, et al. Viral load, genotypes, and mutants in hepatitis B virus-related hepatocellular carcinoma: special emphasis on patients with early hepatocellular carcinoma. Dig Dis Sci 2012;57:232-8. [DOI] [PubMed] [Google Scholar]
- 52.Yan T, Li K, Li F, et al. T1846 and A/G1913 are associated with acute on chronic liver failure in patients infected with hepatitis B virus genotypes B and C. J Med Virol 2011;83:996-1004. [DOI] [PubMed] [Google Scholar]
- 53.Tatsukawa M, Takaki A, Shiraha H, et al. Hepatitis B virus core promoter mutations G1613A and C1653T are significantly associated with hepatocellular carcinoma in genotype C HBV-infected patients. BMC Cancer 2011;11:458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu S, Xie J, Yin J, et al. A matched case-control study of hepatitis B virus mutations in the preS and core promoter regions associated independently with hepatocellular carcinoma. J Med Virol 2011;83:45-53. [DOI] [PubMed] [Google Scholar]
- 55.Bai X, Zhu Y, Jin Y, et al. Temporal acquisition of sequential mutations in the enhancer II and basal core promoter of HBV in individuals at high risk for hepatocellular carcinoma. Carcinogenesis 2011;32:63-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Welschinger R, Kew MC, Viana R, et al. T1653 mutation in the enhancer II region of the hepatitis B virus genome in southern African Blacks with hepatocellular carcinoma. Eur J Gastroenterol Hepatol 2010;22:541-5. [DOI] [PubMed] [Google Scholar]
- 57.Jin Y, Zhu Y, Chen TY, et al. The association of hepatitis B virus genotype and the basal core promoter mutation in Qidong, China. Zhonghua Gan Zang Bing Za Zhi 2010;18:511-5. [DOI] [PubMed] [Google Scholar]
- 58.Asim M, Malik A, Sarma MP, et al. Hepatitis B virus BCP, Precore/core, X gene mutations/genotypes and the risk of hepatocellular carcinoma in India. J Med Virol 2010;82:1115-25. [DOI] [PubMed] [Google Scholar]
- 59.Kim JK, Chang HY, Lee JM, et al. Specific mutations in the enhancer II/core promoter/precore regions of hepatitis B virus subgenotype C2 in Korean patients with hepatocellular carcinoma. J Med Virol 2009;81:1002-8. [DOI] [PubMed] [Google Scholar]
- 60.Abe K, Thung SN, Wu HC, et al. Pre-S2 deletion mutants of hepatitis B virus could have an important role in hepatocarcinogenesis in Asian children. Cancer Sci 2009;100:2249-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dong Q, Chan HL, Liu Z, et al. A1762T/G1764A mutations of hepatitis B virus, associated with the increased risk of hepatocellular carcinoma, reduce basal core promoter activities. Biochem Biophys Res Commun 2008;374:773-6. [DOI] [PubMed] [Google Scholar]
- 62.Chen GG, Li MY, Ho RL, et al. Identification of hepatitis B virus X gene mutation in Hong Kong patients with hepatocellular carcinoma. J Clin Virol 2005;34:7-12. [DOI] [PubMed] [Google Scholar]
- 63.Yuen MF, Tanaka Y, Mizokami M, et al. Role of hepatitis B virus genotypes Ba and C, core promoter and precore mutations on hepatocellular carcinoma: a case control study. Carcinogenesis 2004;25:1593-8. [DOI] [PubMed] [Google Scholar]
- 64.Kuang SY, Jackson PE, Wang JB, et al. Specific mutations of hepatitis B virus in plasma predict liver cancer development. Proc Natl Acad Sci U S A 2004;101:3575-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang XH, Wang PL. Relationship between mutation on precore region of integrated HBV DNA and p53 gene mutation in hepatocellular carcinoma. Ai Zheng (in Chinese) 2003;22:715-8. [PubMed] [Google Scholar]
- 66.Bläckberg J, Kidd-Ljunggren K. Mutations within the hepatitis B virus genome among chronic hepatitis B patients with hepatocellular carcinoma. J Med Virol 2003;71:18-23. [DOI] [PubMed] [Google Scholar]
- 67.Chen WN, Oon CJ. Mutations and deletions in core promoter and precore stop codon in relation to viral replication and liver damage in Singaporean hepatitis B virus carriers. Eur J Clin Invest 2000;30:787-92. [DOI] [PubMed] [Google Scholar]
- 68.Mathews P, Lee D, Chung YH, et al. Effects of genomic changes in hepatitis B virus on postoperative recurrence and survival in patients with hepatocellular carcinoma. Ann Surg Oncol 2013;20:1216-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heo NY, Lee HC, Park YK, et al. Lack of association between hepatitis B virus pre-S mutations and recurrence after surgical resection in hepatocellular carcinoma. J Med Virol 2013;85:589-96. [DOI] [PubMed] [Google Scholar]
- 70.Tsai HW, Lin YJ, Lin PW, et al. A clustered ground-glass hepatocyte pattern represents a new prognostic marker for the recurrence of hepatocellular carcinoma after surgery. Cancer 2011;117:2951-60. [DOI] [PubMed] [Google Scholar]
- 71.Ryu HJ, Kim do Y, Park JY, et al. Clinical features and prognosis of hepatocellular carcinoma with respect to pre-S deletion and basal core promoter mutations of hepatitis B virus Genotype C2. J Med Virol 2011;83:2088-95. [DOI] [PubMed] [Google Scholar]
- 72.Yeh CT, So M, Ng J, et al. Hepatitis B virus-DNA level and basal core promoter A1762T/G1764A mutation in liver tissue independently predict postoperative survival in hepatocellular carcinoma. Hepatology 2010;52:1922-33. [DOI] [PubMed] [Google Scholar]
- 73.Yeung P, Wong DK, Lai CL, et al. Association of hepatitis B virus pre-S deletions with the development of hepatocellular carcinoma in chronic hepatitis B. J Infect Dis 2011;203:646-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qu L, Kuai X, Liu T, et al. Pre-S deletion and complex mutations of hepatitis B virus related to young age hepatocellular carcinoma in Qidong, China. PLoS One 2013;8:e59583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qu LS, Liu TT, Jin F, et al. Combined pre-S deletion and core promoter mutations related to hepatocellular carcinoma: A nested case-control study in China. Hepatol Res 2011;41:54-63. [DOI] [PubMed] [Google Scholar]
- 76.Wang XY, Li GJ, Dong BQ, et al. A matched nested case-control study of the relationship between hepatitis B virus pre-S deletion mutations and hepatocellular carcinoma. Ying Yong Yu Fang Yi Xue (in Chinese) 2010;16:5-8. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The reference lists of the 36 excluded studies. Studies were excluded because: (I) studies (41-50) did not use a prospective design (n=27) (51-67); (II) studies reported HCC survival or prognosis as the sole outcome of interest (n=5) (68-72); (III) no relative risks (RRs) or 95% confidence intervals (CIs) were reported, or there were not enough data to calculate them (n=1) (73); or (IV) newer data were available (n=3) (74-76).