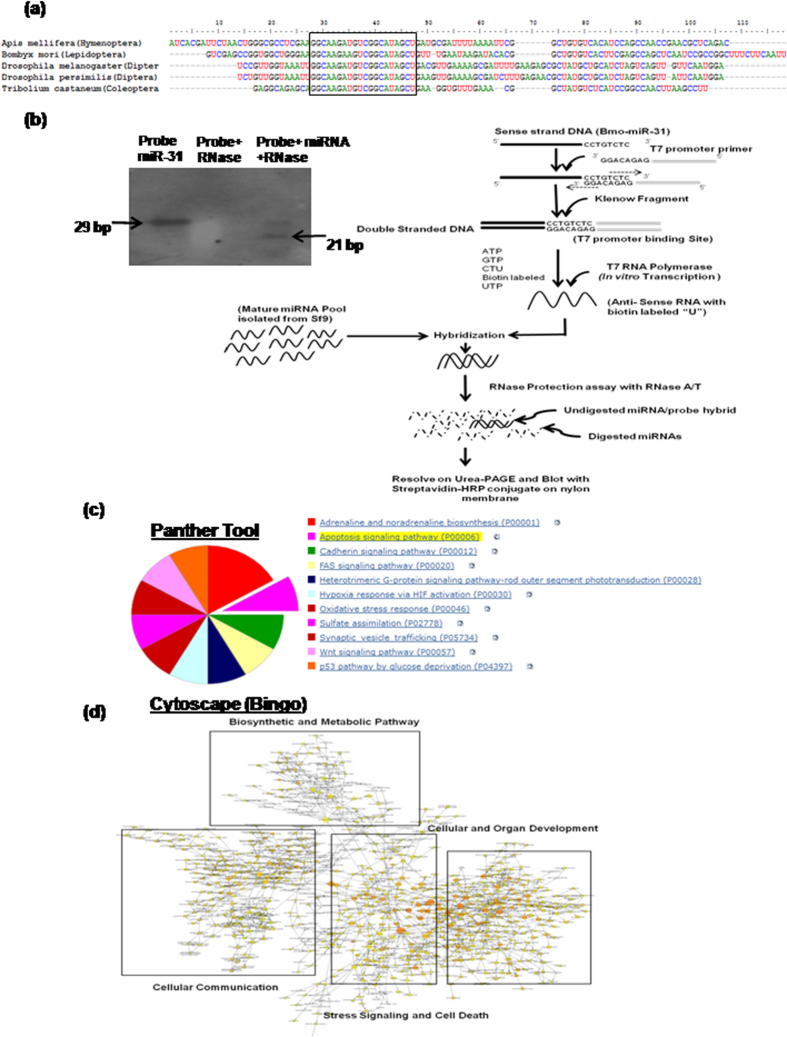
Figure 1. Identification and functional annotation of miRNA-31 in Sf9 cells.
(a) Clustal W analysis of miR-31 sequences from five different species of the same order shows miR-31 conservation at mature miRNA level; (b) As shown in the schematic, in vitro transcribed biotinylated probes were used with miRNAs isolated from Sf9 cells for RNase protection assay followed by chemiluminescent blotting (with streptavidin- HRP conjugate) for the identification of miR-31. First lane on the left shows the in vitro transcribed probe. Middle lane shows the probe digested with RNase. In the right lane, upper band shows undigested probe and lower band depicts 21 bp mature miR-31; (c) Sf-miR-31 sequence was queried against all the predicted UTRs of Bombyx mori and the predicted potential targets were mapped using panther tool for analysis of molecular functions and biological processes. Predicted targets of Sf-miR-31 majorly include regulatory pathways of apoptotic signaling, among few other important cellular functions. (d) Cytoscape (Bingo) tool was used for Gene Ontology term clustering of all the predicted targets. The four major biological pathways predicted as regulated by Sf-miR-31 are shown in different groups within the hollow boxes.