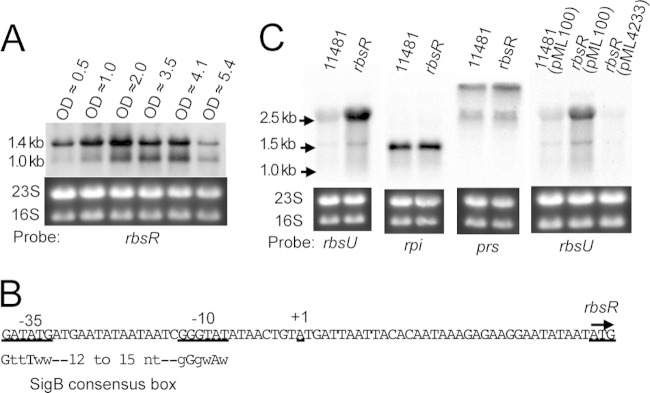
FIG 5.
Transcription analyses. Total RNA (5 μg) from each sample used for Northern blotting was denatured in formamide, applied to TBE-agarose gels, and hybridized with the Dig-labeled specific probe indicated below each blot. (A) Expression of rbsR in CYL11481 cultures grown to different OD600s, as indicated. (B) Map of TSS and predicted promoter of rbsR. The SigB binding consensus is shown below, with capital letters denoting highly conserved nucleotides and lowercase letters denoting poorly conserved nucleotides (w = A or T). (C) Effects of RbsR on rbsU, rpi, and prs transcription. Total RNAs were prepared from 4-h cultures (OD600, ∼4.0) of S. aureus CYL11481 or the rbsR mutant (CYL12834) and hybridized with specific probes, as indicated. The expression of rbsU was also assessed by complementation of rbsR by using pML100-rbsR (pML4233) in the presence of ATc.