Abstract
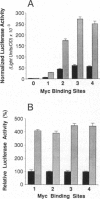
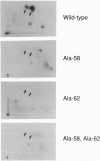
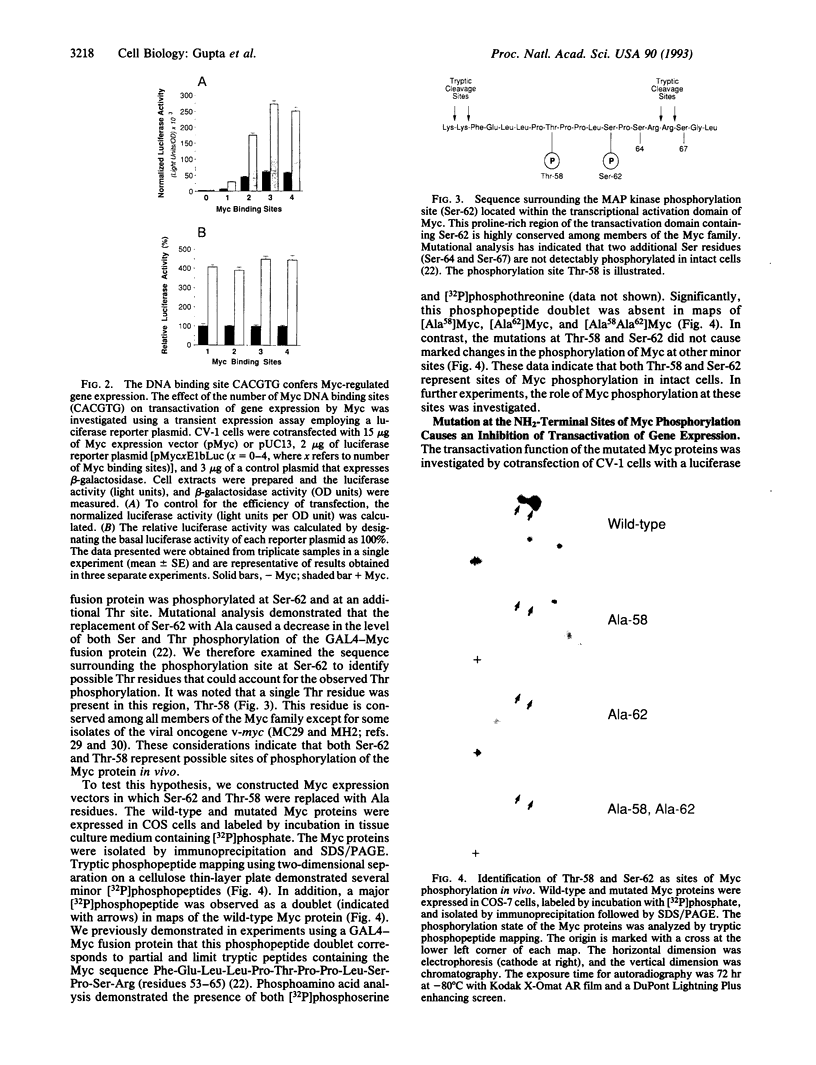
The product of the human c-myc protooncogene (Myc) is a sequence-specific DNA binding protein. Here, we demonstrate that the placement of the specific Myc DNA binding site CACGTG upstream of a luciferase reporter gene conferred Myc-stimulated expression that was inhibited by the overexpression of the basic-helix-loop-helix/leucine zipper protein Max. It was observed that Myc was phosphorylated in vivo within the NH2-terminal domain at Thr-58 and Ser-62. Replacement of these phosphorylation sites with Ala residues caused a marked decrease in Myc-stimulated reporter gene expression. In contrast, the replacement of Thr-58 or Ser-62 with an acidic residue (Glu) caused only a small inhibition of transactivation. Together, these data demonstrate that the NH2-terminal phosphorylation sites Thr-58 and Ser-62 are required for high levels of transactivation of gene expression by Myc.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alvarez E., Northwood I. C., Gonzalez F. A., Latour D. A., Seth A., Abate C., Curran T., Davis R. J. Pro-Leu-Ser/Thr-Pro is a consensus primary sequence for substrate protein phosphorylation. Characterization of the phosphorylation of c-myc and c-jun proteins by an epidermal growth factor receptor threonine 669 protein kinase. J Biol Chem. 1991 Aug 15;266(23):15277–15285. [PubMed] [Google Scholar]
- Andersson S., Davis D. L., Dahlbäck H., Jörnvall H., Russell D. W. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989 May 15;264(14):8222–8229. [PubMed] [Google Scholar]
- Barrett J., Birrer M. J., Kato G. J., Dosaka-Akita H., Dang C. V. Activation domains of L-Myc and c-Myc determine their transforming potencies in rat embryo cells. Mol Cell Biol. 1992 Jul;12(7):3130–3137. doi: 10.1128/mcb.12.7.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binétruy B., Smeal T., Karin M. Ha-Ras augments c-Jun activity and stimulates phosphorylation of its activation domain. Nature. 1991 May 9;351(6322):122–127. doi: 10.1038/351122a0. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Kretzner L., Blackwood E. M., Eisenman R. N., Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990 Nov 23;250(4984):1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Eisenman R. N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991 Mar 8;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Kretzner L., Eisenman R. N. Myc and Max function as a nucleoprotein complex. Curr Opin Genet Dev. 1992 Apr;2(2):227–235. doi: 10.1016/s0959-437x(05)80278-3. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Lüscher B., Eisenman R. N. Myc and Max associate in vivo. Genes Dev. 1992 Jan;6(1):71–80. doi: 10.1101/gad.6.1.71. [DOI] [PubMed] [Google Scholar]
- Chen R. H., Sarnecki C., Blenis J. Nuclear localization and regulation of erk- and rsk-encoded protein kinases. Mol Cell Biol. 1992 Mar;12(3):915–927. doi: 10.1128/mcb.12.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M. D. The myc oncogene: its role in transformation and differentiation. Annu Rev Genet. 1986;20:361–384. doi: 10.1146/annurev.ge.20.120186.002045. [DOI] [PubMed] [Google Scholar]
- Dang C. V., Barrett J., Villa-Garcia M., Resar L. M., Kato G. J., Fearon E. R. Intracellular leucine zipper interactions suggest c-Myc hetero-oligomerization. Mol Cell Biol. 1991 Feb;11(2):954–962. doi: 10.1128/mcb.11.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M., Schirm S., Bishop J. M. The MYC protein activates transcription of the alpha-prothymosin gene. EMBO J. 1991 Jan;10(1):133–141. doi: 10.1002/j.1460-2075.1991.tb07929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F. A., Raden D. L., Davis R. J. Identification of substrate recognition determinants for human ERK1 and ERK2 protein kinases. J Biol Chem. 1991 Nov 25;266(33):22159–22163. [PubMed] [Google Scholar]
- Gonzalez G. A., Montminy M. R. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989 Nov 17;59(4):675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- Halazonetis T. D., Kandil A. N. Determination of the c-MYC DNA-binding site. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6162–6166. doi: 10.1073/pnas.88.14.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann S. R., Abrams H. D., Rohrschneider L. R., Eisenman R. N. Proteins encoded by v-myc and c-myc oncogenes: identification and localization in acute leukemia virus transformants and bursal lymphoma cell lines. Cell. 1983 Oct;34(3):789–798. doi: 10.1016/0092-8674(83)90535-4. [DOI] [PubMed] [Google Scholar]
- Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989 Apr 15;77(1):51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Hunter T., Karin M. The regulation of transcription by phosphorylation. Cell. 1992 Aug 7;70(3):375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- Håvarstein L. S., Morgan I. M., Wong W. Y., Vogt P. K. Mutations in the Jun delta region suggest an inverse correlation between transformation and transcriptional activation. Proc Natl Acad Sci U S A. 1992 Jan 15;89(2):618–622. doi: 10.1073/pnas.89.2.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan N. C., Flordellis C. S., Mark G. E., Duesberg P. H., Papas T. S. Nucleotide sequence of avian carcinoma virus MH2: two potential onc genes, one related to avian virus MC29 and the other related to murine sarcoma virus 3611. Proc Natl Acad Sci U S A. 1984 May;81(10):3000–3004. doi: 10.1073/pnas.81.10.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato G. J., Barrett J., Villa-Garcia M., Dang C. V. An amino-terminal c-myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990 Nov;10(11):5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato G. J., Lee W. M., Chen L. L., Dang C. V. Max: functional domains and interaction with c-Myc. Genes Dev. 1992 Jan;6(1):81–92. doi: 10.1101/gad.6.1.81. [DOI] [PubMed] [Google Scholar]
- Kelly K., Cochran B. H., Stiles C. D., Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983 Dec;35(3 Pt 2):603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- Kerkhoff E., Bister K., Klempnauer K. H. Sequence-specific DNA binding by Myc proteins. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4323–4327. doi: 10.1073/pnas.88.10.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher B., Kuenzel E. A., Krebs E. G., Eisenman R. N. Myc oncoproteins are phosphorylated by casein kinase II. EMBO J. 1989 Apr;8(4):1111–1119. doi: 10.1002/j.1460-2075.1989.tb03481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkelä T. P., Koskinen P. J., Västrik I., Alitalo K. Alternative forms of Max as enhancers or suppressors of Myc-ras cotransformation. Science. 1992 Apr 17;256(5055):373–377. doi: 10.1126/science.256.5055.373. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Lawe D., Ziff E. B. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991 May 3;65(3):395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Ziff E. B. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991 Jan 11;251(4990):186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- Pulverer B. J., Kyriakis J. M., Avruch J., Nikolakaki E., Woodgett J. R. Phosphorylation of c-jun mediated by MAP kinases. Nature. 1991 Oct 17;353(6345):670–674. doi: 10.1038/353670a0. [DOI] [PubMed] [Google Scholar]
- Reddy E. P., Reynolds R. K., Watson D. K., Schultz R. A., Lautenberger J., Papas T. S. Nucleotide sequence analysis of the proviral genome of avian myelocytomatosis virus (MC29). Proc Natl Acad Sci U S A. 1983 May;80(9):2500–2504. doi: 10.1073/pnas.80.9.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach P. J. Multisite and hierarchal protein phosphorylation. J Biol Chem. 1991 Aug 5;266(22):14139–14142. [PubMed] [Google Scholar]
- Saksela K., Mäkelä T. P., Hughes K., Woodgett J. R., Alitalo K. Activation of protein kinase C increases phosphorylation of the L-myc trans-activator domain at a GSK-3 target site. Oncogene. 1992 Feb;7(2):347–353. [PubMed] [Google Scholar]
- Seth A., Alvarez E., Gupta S., Davis R. J. A phosphorylation site located in the NH2-terminal domain of c-Myc increases transactivation of gene expression. J Biol Chem. 1991 Dec 15;266(35):23521–23524. [PubMed] [Google Scholar]
- Seth A., Gonzalez F. A., Gupta S., Raden D. L., Davis R. J. Signal transduction within the nucleus by mitogen-activated protein kinase. J Biol Chem. 1992 Dec 5;267(34):24796–24804. [PubMed] [Google Scholar]
- Smeal T., Binetruy B., Mercola D. A., Birrer M., Karin M. Oncogenic and transcriptional cooperation with Ha-Ras requires phosphorylation of c-Jun on serines 63 and 73. Nature. 1991 Dec 12;354(6353):494–496. doi: 10.1038/354494a0. [DOI] [PubMed] [Google Scholar]
- Stone J., de Lange T., Ramsay G., Jakobovits E., Bishop J. M., Varmus H., Lee W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987 May;7(5):1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgett J. R. A common denominator linking glycogen metabolism, nuclear oncogenes and development. Trends Biochem Sci. 1991 May;16(5):177–181. doi: 10.1016/0968-0004(91)90071-3. [DOI] [PubMed] [Google Scholar]