Figure 4.
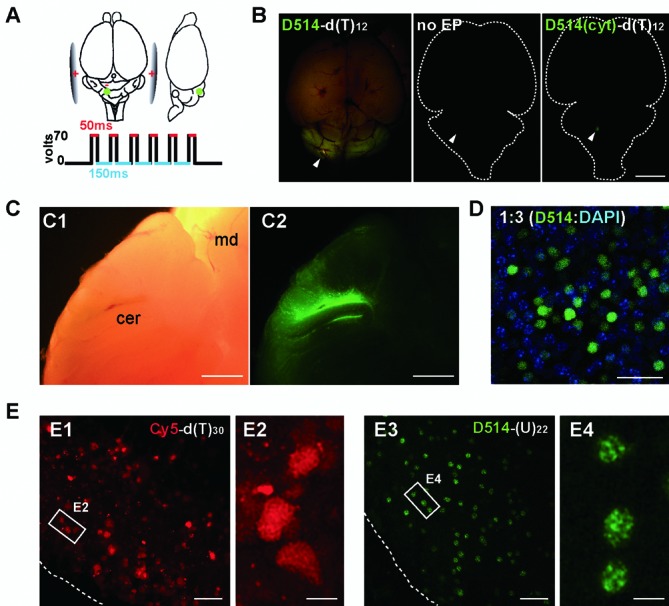
In vivo labeling of target RNAs in mouse brains. (A) Illustration of the in vivo electroporation procedure for delivering ECHO probes into the cerebellum of an anesthetized mouse. Green dots: probe injection sites (500 μm in depth from the surface). (B) Dorsal fluorescence view of P7 mice brains electroporated with D514-d(T)12 (left), injected with D514-d(T)12 with no current delivery (middle), electroporated with a single-mismatched probe that contains cytidine in place of the central thymidine (right). White arrowheads are pointed to the injection sites. (C) Bright-field (C1) and fluorescence (C2) views in sagittally partitioned brain halves. cer, cerebellum; md, midbrain. (D) Confocal images (LSM780) of permeabilized cerebellar slices stained with DAPI (blue) after in vivo electroporation of ECHO probe (green). One out of every three cells in the affected regions contained D514 fluorescence, indicating that delivery efficiency is roughly 30%. DAPI: 405 nm excitation, 410–500 nm detection; D514: 514 nm excitation, 517–597 nm detection; 0.132 × 0.132 μm pixel size; 0.64 μs pixel dwell times. Image stacks of 1.36 μm optical slices at 0.68 μm interval up to 8.9 μm depth. (E) Acute cerebellar slices were prepared after in vivo electroporation and imaged using a standard confocal laser scanning setup (FV1000). Electroporation of oligonucleotide probes labeled with a conventional dye Cy5-d(T)30 showed high fluorescence background at both intracellular and extracellular locations (E1). No distinguishable intranuclear structures were resolved by Cy5-d(T)30 labeling (E2). In contrast, D514-(U)22 reveals robust fluorescence in nuclei with relatively low background (E3); At higher magnification, D514-(U)22 reveals readily distinguishable poly(A) nuclear speckles in individual cerebellar cells (E4). Scale bars: 5 mm (B), 1 mm (C), 20 μm (D, E1, 2), 5 μm (E3, 4). D514: 515 nm excitation, 530–575 nm detection; Cy5: 635 nm excitation, 655–755 nm detection;. 0.207× 0.207 μm pixel size; 8 μs pixel dwell times.
