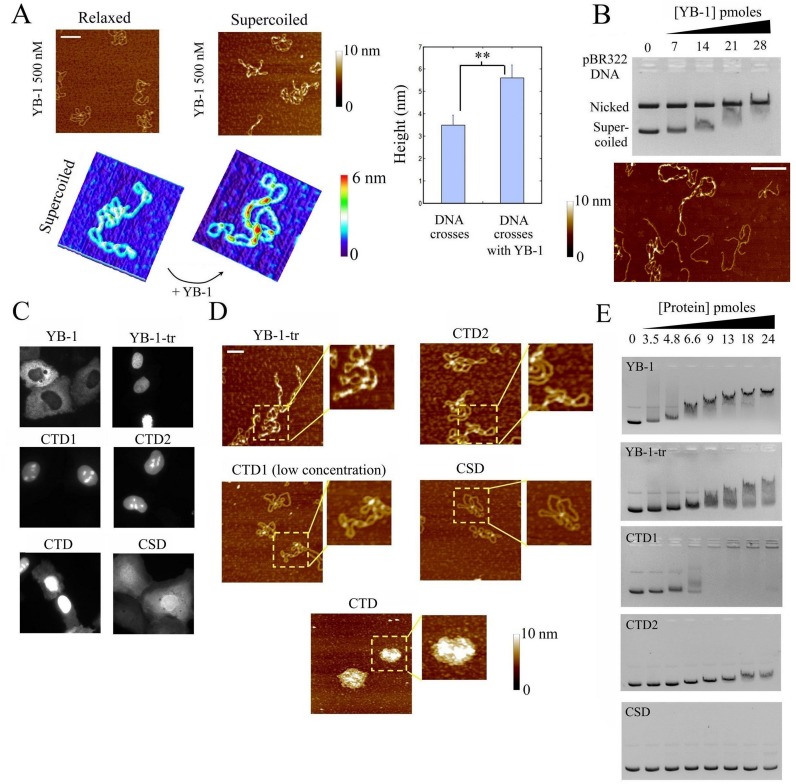
Figure 5.
YB-1 binds preferentially supercoiled DNA at crosses (A) Left panel: The interaction of YB-1 (500 nM) with nicked or supercoiled pBR322 DNA (2 nM) was probed by AFM on mica. YB-1 forms short multimers on supercoiled pBR322 DNA, especially at DNA crosses, while the binding of YB-1 to relaxed plasmids was not detected. Scale bar: 200 nm. Right panel: Analysis of the heights at DNA crosses of supercoiled DNA in the presence or absence of YB-1. The significant increase in height of DNA crosses in the presence of YB-1 reveals its multimerization at crosses. (B) Upper panel: Gel mobility shift assay probing the binding of YB-1 to an equimolar mixture of supercoiled and nicked pBR322 DNA (70 fmoles each). A significant decrease in the mobility of supercoiled DNA occurs at lower concentration than for relaxed plasmid. Two non-exclusive explanations can be advanced for this: (i) YB-1 binds to both supercoiled and relaxed DNA but only reduces the mobility of supercoiled DNA by modifying its conformation, (ii) YB-1 preferentially binds to supercoiled DNA. AFM data (see lower panel) indicate that YB-1 indeed preferentially binds to DNA crosses. Lower panel: AFM image of linearized and supercoiled pBR322 DNA (1 nM each) after their incubation in the presence of 500 nM YB-1. We observed that YB-1 preferentially binds to supercoiled DNA at DNA crosses in the presence of linear DNA. Scale bar: 150 nm. (C) Cellular location of the indicated GFP-labeled YB-1 constructs after their expression in normal rat kidney cells. YB-1-tr, CTD, CTD1, CTD2 were rather located in the nucleus in contrast with the cytoplasmic location of YB-1 and the homogenous distribution of CSD. (D) The presence of the indicated YB-1 constructs on supercoiled pBR322 DNA (2 nM) was probed by AFM. YB-1-tr (500 nM) form short multimers at DNA crosses in contrast with CTD1 (100 nM) and CTD2 (500 nM). CTD (500 nM) and CTD1 (500 nM, not shown) lead to the formation of mRNA-containing granules. In the presence of CSD, no interaction with supercoiled DNA was observed. Scale bar: 100 nm. (E) Gel mobility shift assay representing the binding of the indicated YB-1 constructs with supercoiled pBR322 DNA concentration. In agreement with the AFM results (D), a significant shift was detected with YB-1-tr. The shift is less marked for CTD2 and for CTD1 than YB-1 and YB-1-tr, before aggregation takes place. No shift was detected with CSD in the conditions explored here.