Figure 1.
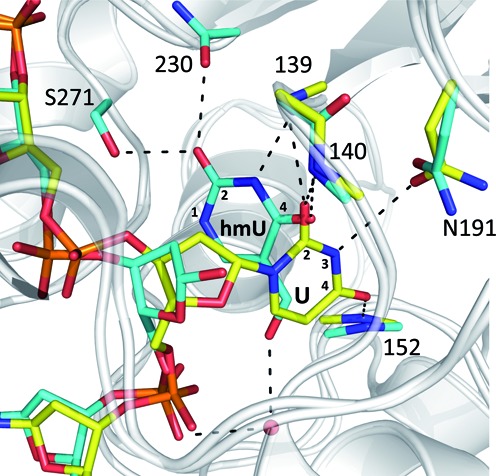
Alignment of two previous structures for TDGcat. A structure of the enzyme–product (E·P) complex for TDGcat processing of a G·hmU mispair is shown with DNA and interacting enzyme moieties colored in cyan (PDB ID: 4FNC) (25). Aligned with this is a structure of the enzyme–substrate (E·S) complex for a G·UF mispair, where UF is a dU analog that flips but is not cleaved by TDG, with DNA and enzyme moieties colored yellow (PDB ID: 3UFJ). For the reported product complex, the putative excised hmU base is markedly displaced from its expected position prior to C-N bond cleavage, forming different contacts with TDG compared to those expected prior to bond cleavage (as indicated by U contacts in the E·S complex). Labels for side chains include the residue type; those for backbone groups include residue number only. Relevant positions of hmU and U are indicated.
