Figure 3.
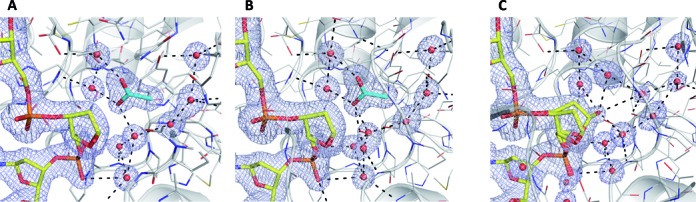
New structures demonstrate that the excised base is absent from enzyme–product complexes resulting from TDGcat action on various substrates. (A) Close-up view of the active site for the enzyme–product (E·P) complex resulting from TDGcat action on a G·hmU substrate, solved at 1.72 Å (PDB ID: 4XEG). The abasic sugar is flipped into the active site, with C1’-OH pointing toward the viewer. The excised base is clearly absent. Coloring is by element, with carbon atoms of the DNA in yellow, the enzyme in white and the acetate in cyan (O, N and P atoms are red, blue, and orange, respectively). Red spheres are water molecules. The 2Fo-Fc omit map, contoured at 1.0 σ, is light blue. The same coloring scheme is used for the two other panels. (B) Structure of the E·P complex for TDGcat acting on a G·U mispair, solved at 1.45 Å (PDBID: 4Z47). Uracil was present at a concentration of 10 mM in solutions used for sample preparation and all crystallization steps. (C) Structure of the E·P complex generated from TDGcat action on a G·U mispair, solved at 1.75 Å (PDB ID: 4Z3A), obtained from crystals grown in acetate-free conditions.
