Figure 5.
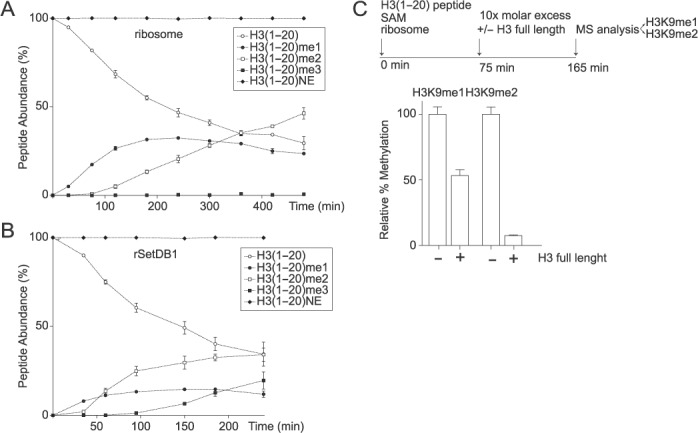
The ribosomal-associated H3K9 HMTase has a non-processive methylation mechanism. Progress curve of histone H3 peptide methylation using unmodified histone H3 peptides from amino acids 1–20 and ribosome complexes (A) or recombinant SetDB1 (B). Each peptide, unmodified, mono-, di- or trimethylated, was quantified and expressed as percent of the total H3 peptide. NE, non-enzymatic control. Standard deviation was obtained from three independent assays. (C) Histone methyltransferase assay using unmodified histone H3 peptides from amino acids 1–20 and ribosome complexes. 75 min after the reaction started, 10X molar excess of histone H3 full length protein was added to the reaction and allowed to proceed for additional 90 min. The methylated peptides (H3K9me1 and H3K9me2) were quantified and expressed as percent of the total H3 peptides. H3K9 HMTase activity of the reaction performed in the presence of competitor was expressed as percent of the reaction performed in the absence of competitor. Standard deviation was obtained from three independent assays.
