Figure 6.
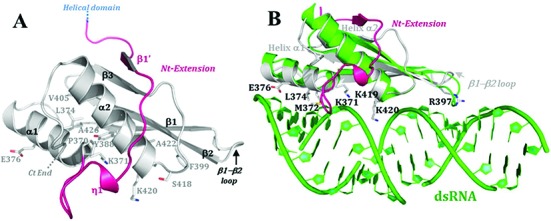
Structure of the dsRBD and model of dsRNA binding mode. (A) Crystal structure of HsDus2dsRBD (Glu347-Lys451) showing all the residues conserved among the dsRBDs family and the novel extension at the N-terminal region of the canonical αβββα fold. (B) Structural alignment of HsDus2dsRBD (in gray) with X. leavis RBPA dsRBD (pdb 1DI2) in complex with a dsRNA fragment (in green). The conserved residues known to be implicated in the dsRNA binding in classical dsRBDs are shown in HsDus2dsRBD. Both dsRBDs align with the exception of the β3 strand and the β1−β2 loop, which are oriented in the opposite direction from dsRNA. In this superposition, several conserved regions of HsDus2dsRBD usually involved in RNA binding in dsRBDs are in contact with the dsRNA without provoking clashes. Therefore, this model could reflect the way HsDus2dsRBD binds a double stranded region of its tRNA substrate. Helix α1 snugly fills the minor groove of the dsRNA, while the N-terminal tip of helix α2 caps the major groove. The basic residues Lys371, Lys419 and Lys420, located in conserved motifs, are oriented towards the phosphodiester backbone of nucleotides from the major groove as in the functional conformations observed in RNAse III (34). Moreover, Arg397 located in the N-terminal region of the β2 strand contacts the minor groove of the dsRNA by interacting with a nucleotide ribose moiety, contrasting with the interaction made by the β1−β2 loop in the RNA binding protein A complex. Although the N-terminal extension carries several basic residues, these do not interact with the dsRNA.
