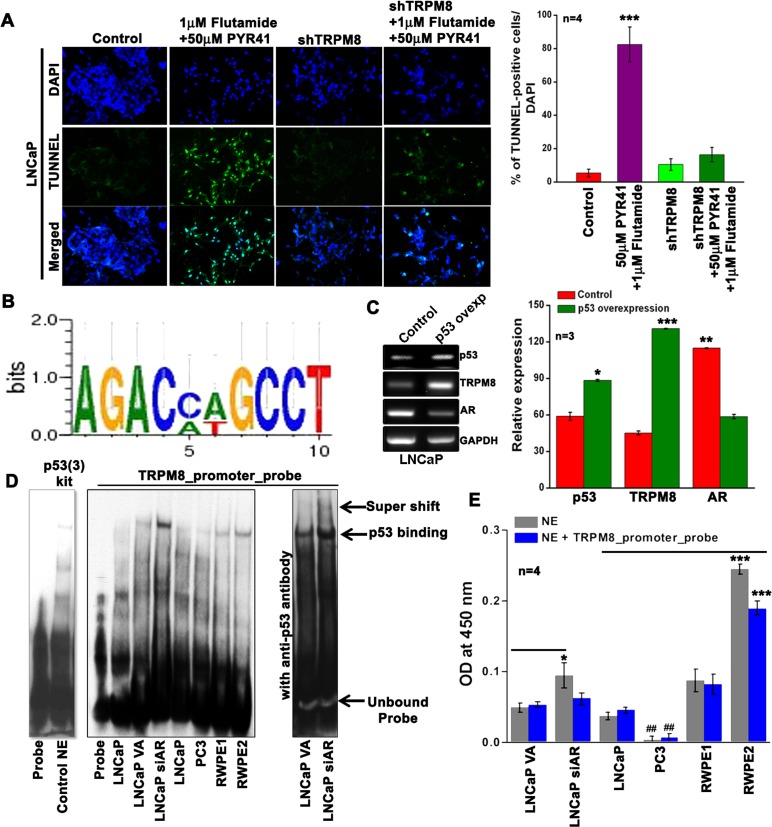
Figure 6. Inverse regulation of TRPM8 by AR and p53.
A. TUNEL staining on LNCaP cells. Green fluorescence represents apoptotic cells and Blue fluorescence represents DAPI. B. The 2064 bp 5′-flanking region of the human trpm8 gene with ARE I and ARE II sites were analyzed further for the presence of possible putative p53 binding sites using TRANSFAC® 7.0. The authenticity of the TFBS was confirmed by other promoter binding software tools; TFBIND, AlGGEN server PROMO (Version 3.0) and Transcription Element Search Software (TESS). The WebLogo 3.4 software was used to generate the sequence logo for p53 binding site (1399 bp upstream of the transcription start site) on trpm8 gene promoter. The error bars indicate an approximate, Bayesian 95% confidence interval. C. Reverse-transcription PCR analysis of control and p53 overexpressing LNCaP cells (primers are listed in Stable 2). D. EMSA was performed with 2 μg of nuclear extract (NE) from control and treated cells. Panel; 1 is for the p53 (3) EMSA probe set in the absence and presence of control NE provided in the kit. Panel; 2 The authenticity of the p53 binding site identified on the TRPM8 promoter was confirmed by performing EMSA using biotinylated probes (STable 3). Panel; 3 shows TRPM8 promoter probe EMSA in the presence of anti-p53 antibody. E. Monitoring p53 activation in the NE using TransAM, assay Kit (Active Motif) in the presence and absence of competing TRPM8 promoter probe. The results are graphically represented as bar diagrams.