Figure 1. Coding somatic variants identified in the 12 pPCL samples.
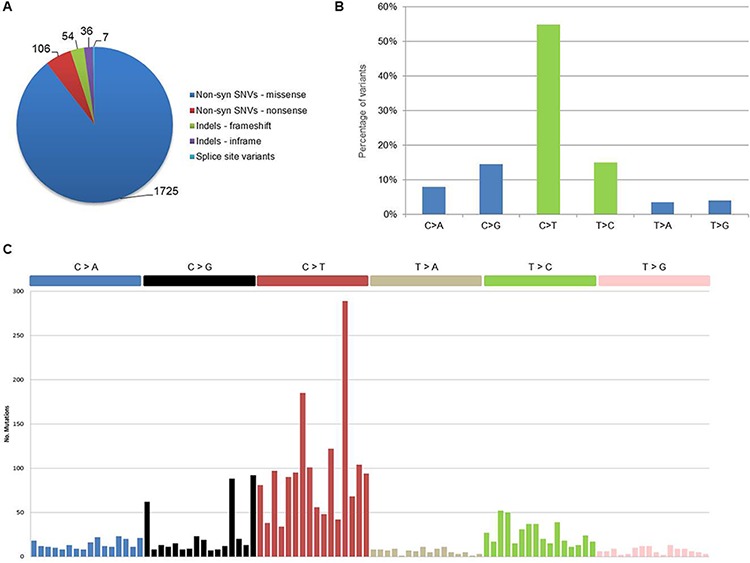
A. Distribution of coding somatic non-silent variants according to amino acid changes or reading frameshifts. B. Classification of coding somatic SNVs (both syn and non-syn) by nucleotide change. Transitions are in green (C > T, T > C) and transversions are in blue (C > A, C > G, T > A, T > G). C. Mutational signatures in pPCL. Coding somatic SNVs identified in the 12 samples were classified according to nucleotide change and sequence context into the 96 possible mutated trinucleotides (listed horizontally according to the conventional order). Substitutions are displayed in different colors along the horizontal axis and number of mutations attributed to each type is shown on the vertical axis.
