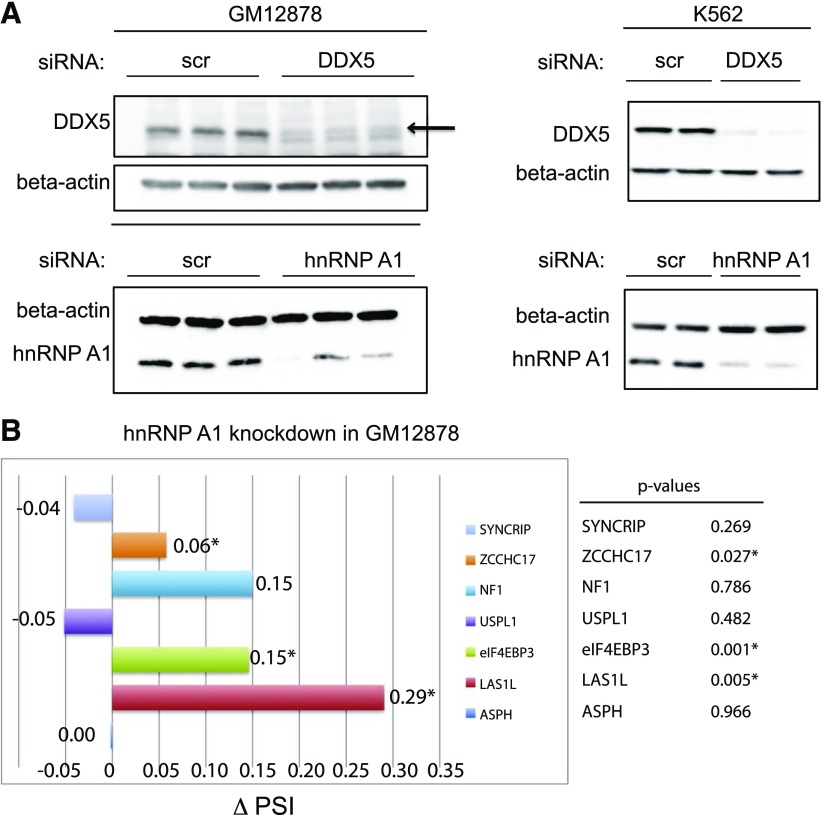
Figure 3.
siRNA-mediated knockdown of hnRNP A1 and DDX5 proteins in GM12878 and K562 cells. A) GM12878 (left) and K562 cells (right) were transfected with nonspecific control siRNA oligos (scr si) or hnRNP A1 or DDX5 siRNA duplexes. After a second round of siRNA transfection, the cells were harvested for preparation of protein lysates, which were subjected to immunoblotting with anti-hnRNP A1 or anti-DDX5 antibodies to assess the efficiency of siRNA-mediated protein knockdown. Immunoblot analysis of the β-actin protein serves as a loading control. B) RT-PCR analysis of several pre-mRNA splicing target mRNAs reveals changes in hnRNP A1-dependent, alternative splicing events, recapitulated by RNAi in GM12878 cells. GM12878 cells were electroporated with nonspecific control siRNAs (scr si) or hnRNP A1 duplex siRNA duplexes. After a 2nd round of siRNA transfection, the cells were harvested for total RNA isolation and subjected to RT-PCR analysis. *P (P < 0.05), calculated by use of a 2-tailed Student's t test, statistically significant. PSI, Percent spliced in.