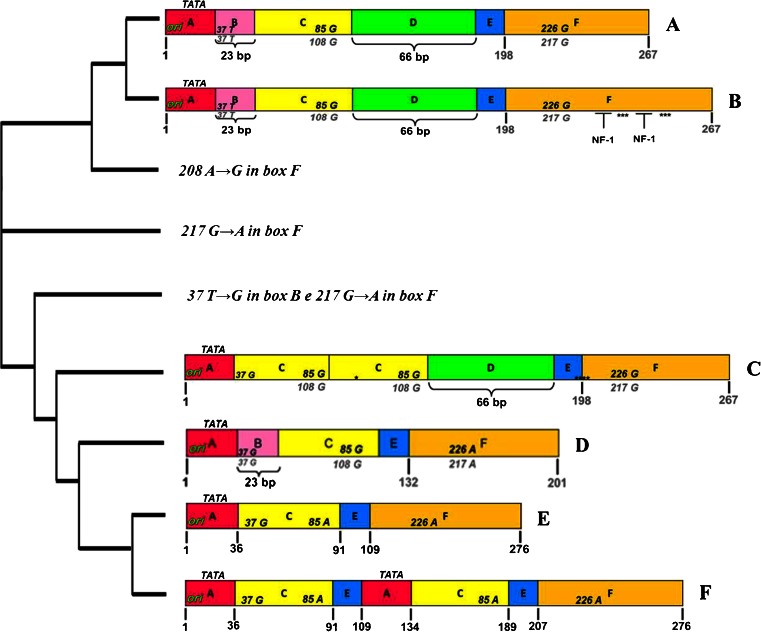
Fig. 3.
Sequences of JCV NCCR found in biological samples of RRMS patients treated with natalizumab. The nucleotide sequences of JCV NCCR were shown from the core of the replication origin (ori) up to the start codon of the gene leader (agnoprotein) of the late viral genes. In A, the nucleotide numbering of the archetype CY sequence, isolated from Yogo and colleagues in 1990, is shown in bold gray. In F, the nucleotide numbering was based on the NCCR sequence of the PML-associated variant Mad-1, sequenced by Frisque and colleagues in 1984, and the nucleotide number is indicated in bold black. In B, the NCCR sequence was characterized by a duplication of cellular transcription factor NF-1 binding site in box F, with loss of the 217 G to A (G → A) nucleotide transition in the same box. It was found in 4 JCV DNA-positive urine samples collected at t0, t1, t2, and t3, respectively, from 1 patient with positive STRATIFY JCV®. In C, a rearranged NCCR was reported, characterized by the deletion of box B with a T to G nucleotide transversion within the cellular transcription factor Spi-B binding site, the duplication of box C, and the presence of box D. It was isolated from PBMC samples of 2 RRMS patients that were STRATIFY JCV® positive after 1 year of treatment with natalizumab (t3). In D, the sequence with an archetype CY-like structural organization was reported but with the deletion of box D and two characteristic point mutations: the 37 T to G (T → G) nucleotide transversion within the binding site for the cellular transcription factor Spi-B in box B and the 217 G to A nucleotide transition in box F. Finally, in E, the sequence model IS is illustrated, consisting of a single sequence from 98 bp, as reported by Jensen and Major JCV NCCR organization (Jensen and Major 2001). The cladogram was performed by software ClustalW2 (Clustal-W2)