Scheme 1.
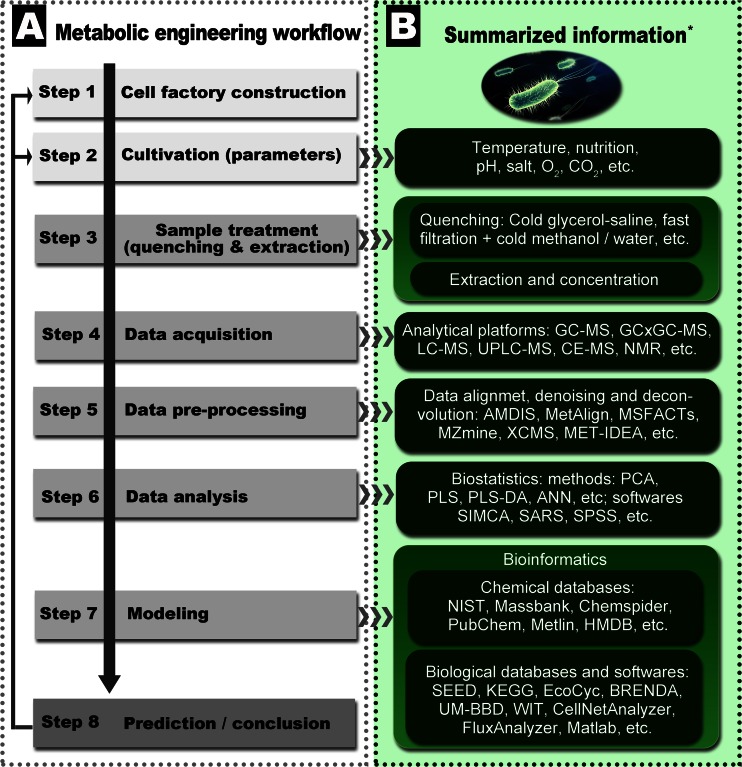
Flowchart and resources for terpenoid microbial metabolomics study. a Microbial metabolic engineering workflow. b Related information of each step for microbial metabolic engineering. * Selected resources: 1. MS data of B. subtilis metabolites (Coulier et al. 2006; Koek et al. 2006; Soga et al. 2003). 2. The metabolomics standards initiative (Fiehn et al. 2007). 3. Microbial metabolomics study examples for terpenoid biosynthesis (Paddon and Keasling 2014; Zhou et al. 2012). 4. Databases, software packages, and protocols (Thiele and Palsson 2010) and http://omictools.com/. 5. Genome-scale data of reconstructed B. subtilis metabolic net (impact of single-gene deletions on growth in B. subtilis) (Oh et al. 2007). 6. Comparative microbial metabolomics study of E. coli, B. subtilis, and S. cerevisiae (van der Werf et al. 2007). 7. The complete genome sequence of B. subtilis (Kunst et al. 1997). 8. Constraint-based modeling methods (Bordbar et al. 2014). 9. Software applications for flux balance analysis (including a software comparative list) (Lakshmanan et al. 2012). 10. Sample treatment methods (Jia et al. 2004; Larsson and Törnkvist 1996; Maharjan and Ferenci 2003; van der Werf et al. 2007; Villas-Bôas and Bruheim 2007)