Figure 2. A strategy for site-specific correction of the sickle cell disease mutation in the HBB locus using homologous recombination stimulated by Cas9 and a guide RNA.
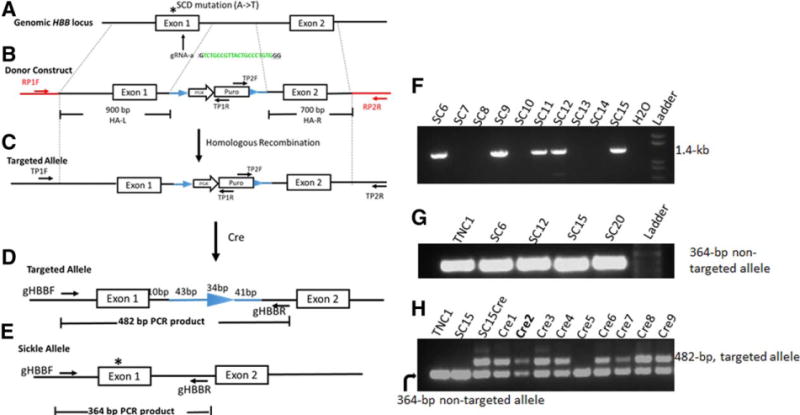
(A) The HBB genomic structure (first two exons), and locations of SCD mutation and gRNA-a targeting HBB exon 1. (B) The gene-targeting donor MGH vector with 2 homology arms (900-bp left arm and 700-bp right arm) introduces a homologous recombination (HR) template for T-to-A replacement in the sickle allele and a loxP-flanked drug-selection cassette PGK-puromycin (puro) to be inserted into the HBB intron 1. (C) Targeted allele following HR with corrected SCD mutation and inserted PGK-puro cassette in intron 1. We used 2 PCR primers (black arrows: TP1F/TP1R and TP2F/TP2R) for initial screening of targeted insertion (TI) indicative of correct HR. (D) Cre-mediated excision removes the PGK-puro cassette and leaves one copy of the loxP DNA (34-bp) and 84-bp linker DNA in HBB intron 1 of the corrected allele. Genomic DNA PCR with primers gHBB-F/gHBB-R produces a 482-bp product from the targeted allele of “Cre” clones. (E) Genomic DNA PCR with primers gHBB-F/gHBB-R produces a 364-bp product from the unintended sickle allele of original and “Cre” clones. (F) PCR screening for targeted integration (TI) by the primer set TP2F/TP2R that detect 3′ TI (1.4-kb). (G) Genomic PCR using the primer set gHBB-F/gHBB-R that amplify the unintended (sickle) allele but not the targeted allele (it is too long to be amplified by PCR with a short extension time) before Cre-mediated excision. (H) PCR screening of Cre-mediated excision of SC15 clones using primer set gHBB-F/gHBB-R. The 482-bp PCR product emerged after Cre-excision as shown in (D).
