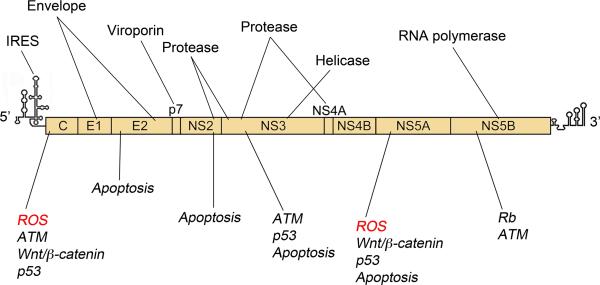
Figure 1. Organization of the 9.7 kb positive-sense HCV RNA genome.
5’ and 3’ untranslated regions (UTRs) contain cis-acting elements essential for virus replication, including a 5’ internal ribosome entry site (IRES) that drives cap-independent expression of a polyprotein (box) that is processed into 10 mature viral proteins by a combination of host and viral proteases. Their functions are highlighted above the genome. The core (C) protein and envelope glycoproteins E1 and E2 are structural components of the virion. Nonstructural (NS) proteins possess functions necessary for replication, including helicase (NS3), protease (NS2 and NS3/4A), and RNA-dependent RNA polymerase (NS5B) activities. NS4B and NS5A drive formation of the ‘membranous web’, a cytoplasmic structure where these proteins accumulate to direct viral RNA synthesis. NS2 and NS5A also function in virion assembly, whereas p7 is essential for egress. Virus-host interactions that may contribute to HCC development are highlighted below the genome. Core and NS5A expression have been linked to the generation of ROS that may contribute to host DNA damage. Multiple HCV proteins interact with and modulate host pathways to facilitate virus replication and may in theory promote carcinogenesis.