Fig. 1.
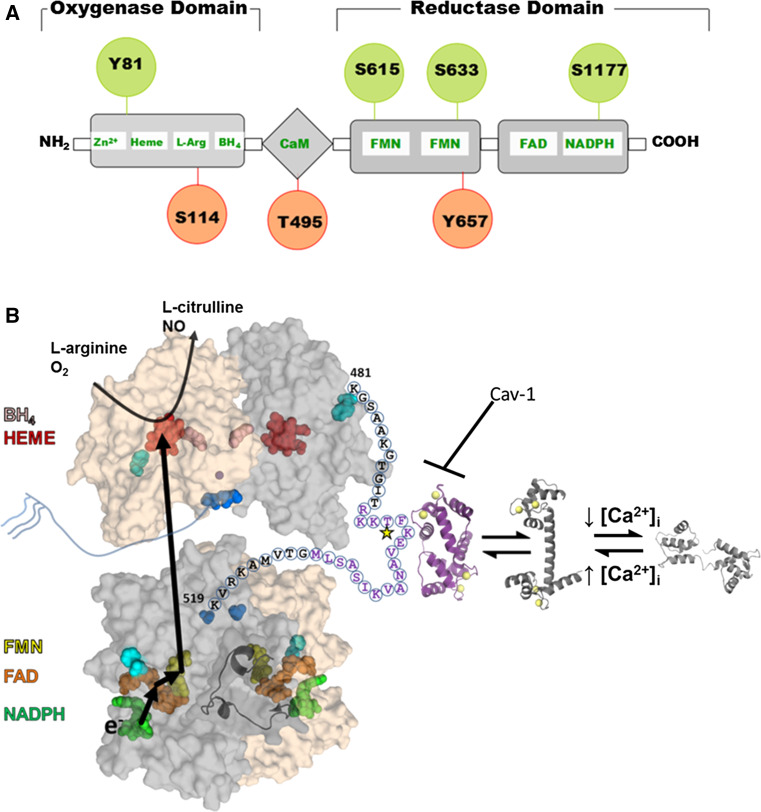
Structure and regulation of eNOS. a eNOS has oxygenase (residues 98–486), Calmodulin (CaM) binding (491–510), and reductase (756–1002) domains. All three domains contain phosphorylation sites (indicated by circles) that modulate eNOS activity (red for decreased activity and green for increased activity). b Structural model of the full length eNOS. The crystal structure of the human oxygenase domain and a homology model of the reductase domain (using the rat nNOS reductase structure (PDBid: 1TLL) as the template) are shown in a surface representation with one subunit colored gray and the other tan. The autoinhibitory loop (residues 596–640) is rendered as a cartoon for one subunit. Cofactors are rendered as spheres and colored: NADPH, green; FAD, orange; FMN, yellow; heme, red; H4biopterin, pink; and zinc, purple. The N-(blue) and C-terminal (cyan) are also rendered as spheres. The linker between the oxygenase and reductase domains is shown for one subunit with the letters indicating the primary sequence. Purple text highlights the CaM binding domain with the star indicating the T495 phosphorylation site. The structure of CaM bound to a peptide corresponding to the eNOS sequence is shown in purple, and the calcium-bound and unbound CaM structures are shown in shades of gray. This model is based on that proposed by Garcin et al. [11]